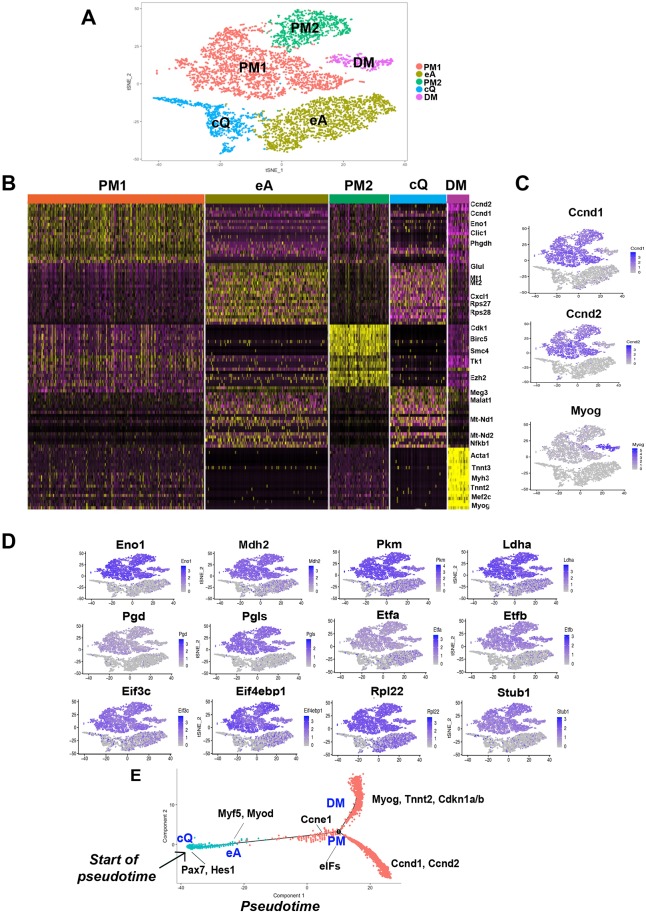
Fig. 3.
Clustering and pseudotemporal trajectories identify transcriptional dynamics of MuSCs and PMs. (A) Graph-based clustering of FACS-isolated MuSCs and PMs showing MuSCs close-to-quiescence (cQ), MuSCs early activation (eA), primary myoblasts cluster 1 (PM1) and cluster 2 (PM2), and differentiating myocytes (DM). (B) Heatmap of the top 20 most variably expressed genes between MuSC cQ, MuSC eA, PM1, PM2 and DM clusters. (C,D) Expression patterns of the cyclin genes Ccnd1, Ccnd2, and myogenin (C) and of glycolytic (Eno1, Pkm, Ldha), mitochondrial (Mdh2, Etfa, Etfb), pentose phosphate (Pgd, Pgls), translation initiation factors (Eif3c, Eif4ebp1), ribosomal (Rpl22) and ubiquitin-ligase (Stub1) genes (D). (E) Pseudotime single cell trajectory reconstructed by Monocle2 for MuSC cQ, MuSC eA and PMs. Pax7 and Hes1 are enriched in MuSC cQ and Myf5 and Myod in MuSC eA. PMs branch off in differentiating myocytes (DM) expressing myogenin (Myog), troponin (Tnnt2) and the cell cycle inhibitors Cdkn1a/b and in PMs that fail to enter differentiation and maintain expression of the cyclins Ccnd1 and Ccdn2.