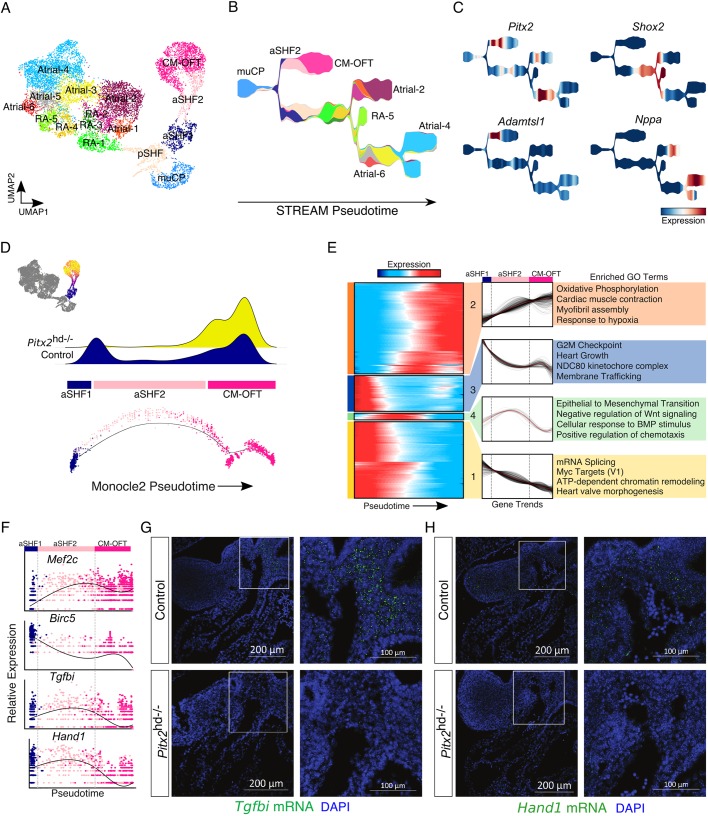
Fig. 4.
Defective cardiac outflow tract differentiation dynamics in Pitx2 null embryonic hearts. (A) UMAP plot showing subclusters derived from merged E10.5 and E13.5 embryonic datasets. (B) STREAM plot showing cluster identities. Colored according to Fig. 4A. (C) STREAM plots showing gene expression. (D) Pseudotime score for each individual outflow tract-associated transcriptome projected onto the UMAP plot (top left). Cells not included in analysis are colored dark gray. Density plot of control and Pitx2 mutant cells across the OFT ontological trajectory (top). Pseudotime minimal spanning tree (MST) plots displaying pseudotime cell identity (bottom). (E) Heatmap showing clusters of dynamic gene expression trends across pseudotime (left). Results of hierarchical clustering of gene expression trends shown in heatmap (middle). Gene ontology (GO) analysis of genes from each indicated dynamic cluster (right). (F) Individual gene expression trends plotted across pseudotime. Cells are colored according to their identity (Fig. 4A). (G) FISH for Tgfbi (green) at E10.5. Nuclei are stained blue (DAPI). (H) FISH for Hand1 (green) at E10.5. Nuclei are stained blue (DAPI). In G and H, images on right are magnifications of the boxed areas in Control (top) and Pitx2 null (bottom) embryos. aSHF, anterior second heart field progenitors; CM-OFT, cardiomyocytes of outflow tract; muCP, progenitors enriched in Pitx2 null tissue; pSHF, posterior second heart field progenitors; RA, right atrial cells.