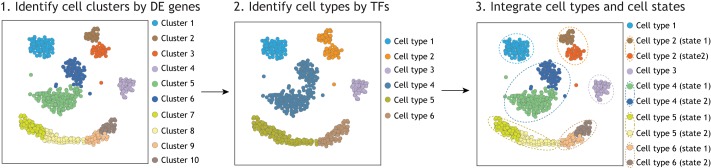
Fig. 2.
A framework for distinguishing cell types from cell states using scRNA-seq. Individual cell transcriptomes can be clustered using typical methods, for example based on DE genes (left), as performed by most scRNA-seq projects. At this point, the cell clusters may not be equivalent to cell types. However, a second step of cell clustering, using only informative transcription factors (TFs), can be used as an additional criterion to distinguish cell types (middle). Thus, by integrating different cell clustering tools (right), we can distinguish cell types from cell states in silico, which can be further tested by molecular and cell biology validation.