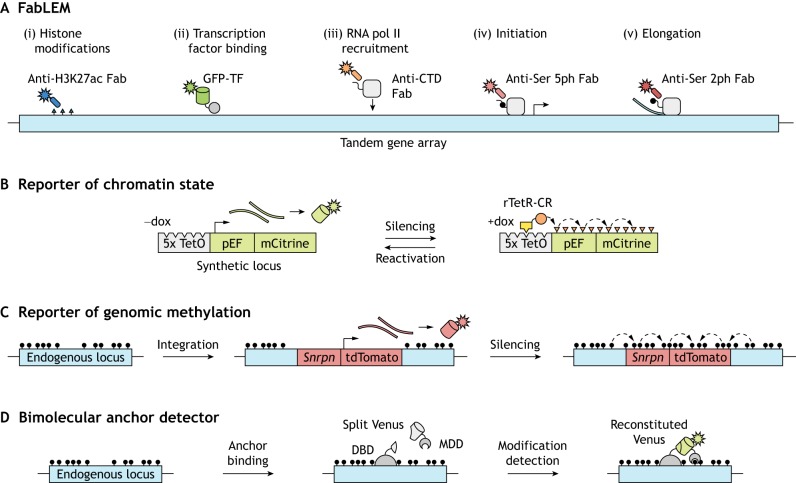
Fig. 4.
Monitoring chromatin modifications and transcription dynamics. (A) Introducing fluorescent antigen-binding fragments (Fabs) into cells for live endogenous modification labeling (LEM) can elucidate chromatin and transcription dynamics to examine their interplay. For example, one can track histone modifications (i), GFP-labeled transcription factors (ii), and phosphorylated serine residues (Ser) in the carboxy-terminal domain (CTD) of RNA pol II that are related to recruitment, initiation and elongation (iii-v). Figure adapted from Stasevich et al. (2014). (B) Gene reporter systems can also be used to follow chromatin dynamics. In the example shown, doxycycline (dox) mediates the recruitment of a chromatin regulator (CR; orange circle) fused to a reverse tetracycline repressor (rTetR; yellow box) to TetO sites upstream of a constitutively expressed fluorescent reporter gene. Recruitment induces gene silencing and deposition of chromatin modifications, and CR release allows gene reactivation. (C) Methylation at an endogenous locus can be monitored by integration of a reporter gene that consists of a minimal imprinted gene promoter (Snrpn) and the coding sequence for a fluorescent protein (tdTomato). DNA methylation spreads from the endogenous locus into the reporter, leading to its silencing. (D) The direct readout of DNA or histone methylation at a locus of interest is possible upon reconstitution of a split fluorescent protein (Venus), the constituents of which are fused to a programmable DNA-binding domain (DBD) and a modification detection domain (MDD).