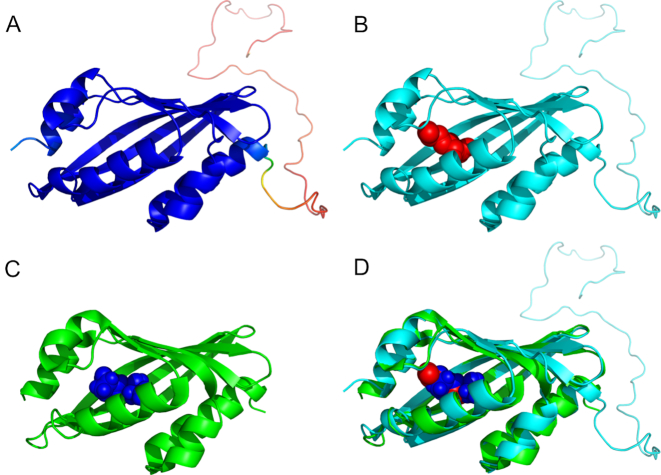
Figure 2.
The IntFOLD5 server predictions for CASP13 target T0971 – comparison of models with the native crystal structure (PDB ID: 6d34). All images were rendered using PyMOL (http://www.pymol.org/). (A) The IntFOLD5 3D model coloured by accuracy self-estimate of local quality using the temperature coloured scheme from blue (indicating residues in the model predicted to be close to the native structure) to red (indicating residues in the model that are far from the native or unstructured). (B) The IntFOLD5 3D model with the main cluster of predicted ligands (red spheres) indicating the predicted location of binding site. (C) The crystal structure of T0971/6d34 with ligand (blue spheres). Note: the disordered domain predicted in the model is absent in the X-ray data. (D) Superposition of the IntFOLD5 model and the native structure.