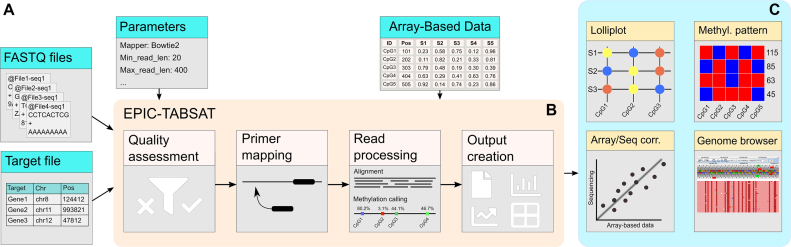
Figure 1.
Workflow of the EPIC-TABSAT software. (A) Input to the software are FASTQ files, a file containing the targets of interest, and a set of parameters. Optionally, an array-based methylation data file can be provided which is combined with the sequencing data. (B) The workflow consists of several quality assessment steps, mapping, methylation calling, and output creation. (C) Methylation calling output and epiallele results along with quality metrics and the correlation of sequencing with the epigenome data are presented.