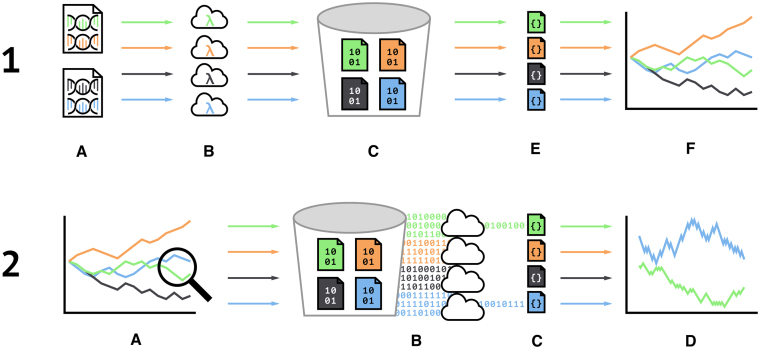
Figure 3.
A diagram displaying the flow of information during initial sequence transformation (1) and sequence querying (2). For initial sequence transformation, FASTA files (1A) are parsed in the user’s browser and submitted asynchronously in parallel to the serverless Lambda functions (1B). If the sequence does not already have an existing transformation using the specified visualization method, the functions transform the sequence and store the data in AWS S3 (1C) in the binary Parquet format. The functions then downsample the transformed data and return it the client’s browser in JSON format (1E) to be plotted (1F). If a user wishes to see more detail of a particular region (2A), the browser sends an asynchronous query containing the location of the region for each plotted DNA sequence to a Lambda function (not shown). In parallel, each function converts the query into a SQL statement and submit the query to S3 Select (2B). S3 Select scans the transformed DNA sequence and returns only the data in the region to the Lambda function, which in turn downsamples to JSON (2C) and returns it to the user’s browser for plotting (2D).