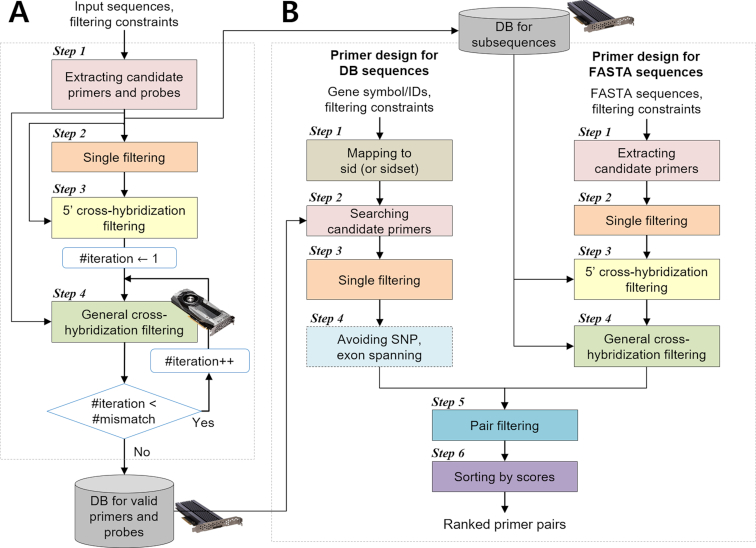
Figure 2.
Overall flow of the MRPrimerW2. MRPrimerW2 consists of two parts, offline processing (A) and online processing (B). (A) Offline processing is independent of user queries and builds two kinds of DBs, ‘DB for valid primers and probes’ and ‘DB for subsequences’, used for online processing. DB for valid primers and probes is used for the tab ‘Primer design for DB sequences’ and DB for subsequences for the tab ‘Primer design for FASTA sequences’. (B) Online processing consists of six steps to design the primers for a given query. It has two different pipelines for two different tabs. In the tab ‘Primer design for DB sequences’, MRPrimerW2 retrieves all candidate primers for a given query and checked filtering constraints on the candidate primers. If a user checks the exon spanning option and/or SNP avoiding option, it evaluates the candidate primers with their exon and/or SNP locus information. In the tab ‘Primer design for FASTA sequences’, MRPrimerW2 extracts all possible subsequences from given FASTA sequence(s) as candidate primers and checks the default filtering constraints for the candidate primers. Then, it performs homology test of the candidate primers using DB for subsequences. Through pair filtering and sorting step, MRPrimerW2 returns the ranked primer pairs.