Figure 6.
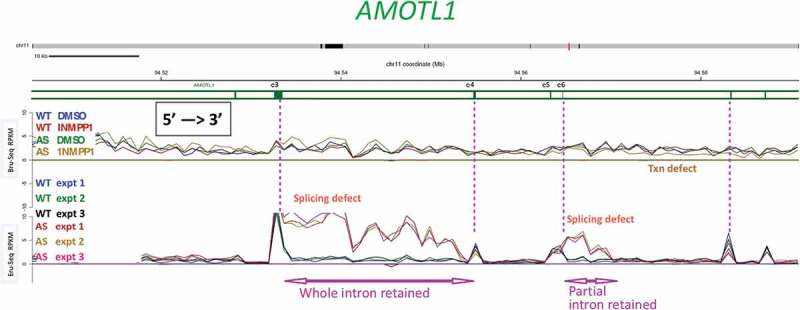
A subset of splice sites is affected by inhibiting CDK12AS for a short time.
Bru-Seq and BruChase-Seq were carried out as described in the text. An internal section of the AMOTL1 gene is shown. The top set of 4 traces (Bru-Seq) show the relative transcription rate (incorporation of BrU in 30 min pulse) in regular HeLa cells (WT) and in CDK12AS HeLa cells in the absence or presence of inhibitory analog 1-NM-PP1. The gold trace (CDK12AS cells + analog) begins to drop below the other three downstream of exon 6 (e6). The bottom set of 6 traces (BruChase-Seq) shows that in ‘WT cells in the presence of analog (blue, green, black), after a 30 min pulse of BrU and a 2 hour chase, exon signals predominate (four exons are indicated by vertical pink dotted lines). The introns are spliced out and degraded (valleys between WT peaks). In contrast, in CDK12AS cells treated identically, the intron following exon 3 (e3) is not spliced out and is “retained.” But the next intron is spliced out normally, and then intronic sequences past exon 6 (e6) are retained. Apparently only part of the intron is retained, since the pulse-chase (BruChase-Seq) traces return to normal about a third of the way through the intron. Farther downstream, splicing looks ~ normal.
