Table 1.
Mediator kinase inhibitors as chemical probes Data listed are for CDK8, but similar results are reported for CDK19 unless otherwise stated. All compounds are reversible inhibitors (i.e. non-covalent) and bind competitively with ATP. Each compound has varying levels of off-target effects and the extent of off-target kinase inhibition (i.e. kinases other than CDK8 or CDK19) was tested more rigorously with some compounds compared with others.
| Name | structure | IC50a | other data | REF |
|---|---|---|---|---|
| BRD6989 | 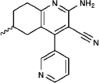 |
500nM in vitro kinase w/CDK8-CCNC |
selective for CDK8 vs. CDK19 | 49 |
| CCT251545 | 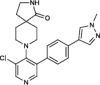 |
5nM in vitro kinase w/ CDK8-CCNC |
Kd = 3.8nM (CDK8-CCNC) IC50 = 65nM (luciferase reporter) PDB: 5BNJ |
122 |
| CCT251921 | 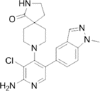 |
2.3nM Lanthascreen or reporter displacement (CDK8-CCNC) |
IC50 ~ 20 nM (luciferase reporter) PDB: 5HBJ |
153 |
| compound 2b | 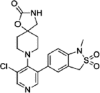 |
1.8nM Lanthascreen or reporter displacement (CDK8-CCNC) |
GI50 = 2nM+ (many lines tested) IC50 ~ 10nM (luciferase reporter) |
123 |
| compound 18 | 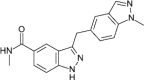 |
10nM Reporter displacement 53nM Lanthascreen (CDK8-CCNC) |
IC50 = 65nM (luciferase reporter) |
154 |
| compound 20 | 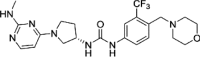 |
17.4nM Lanthascreen (CDK8-CCNC) |
IC50 = 6.5nM (luciferase reporter) PDB: 5HVY |
155 |
| compound 25 | 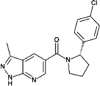 |
2.6nM Lanthascreen or reporter displacement (CDK8-CCNC) |
IC50 = 6.5nM (luciferase reporter) PDB: 5IDN |
156 |
| compound 32 | 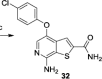 |
1.5nM assay method unclear |
GI50 = 5.4µM* (HCT116) selective for CDK8 vs. CDK19 |
71 |
| compound 51 | 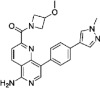 |
5.1nM Lanthascreen or reporter displacement (CDK8-CCNC) |
IC50 = 7.2nM (luciferase reporter) |
157 |
| cortistatin A | 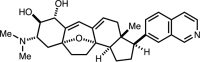 |
12nM in vitro kinase w/ CDK8 module# 100µM ATP |
Kd = 0.2nM (CDK8-CCNC) GI50 = 5nM (MOLM-14) PDB: 4CRL |
48 |
| SEL120-34A | 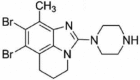 |
4.4nM in vitro kinase w/ CDK8-CCNC 10µM ATP |
Kd = 3nM (CDK8-CCNC) GI50 ~ 12nM (SKNO-1) |
158 |
| senexin A | 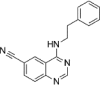 |
280nM in vitro kinase w/ CDK8-CCNC |
Kd ~ 800nM (CDK8-CCNC) |
159 |
| sorafenib | 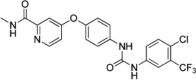 |
32.5nM Lanthascreen (CDK8-CCNC) |
Kd = 100nM (CDK8-CCNC) PDB: 3RGF |
155 160 |
aThe IC50 results determined from kinase assays will be dependent on ATP concentration, with higher [ATP] yielding higher IC50 values. [ATP] used in the assays is noted if reported.
bOther compounds with similar activity were tested in this study.
*In this study, the GI50 was determined to be due to an off-target effect based upon studies in CDK8 and/or CDK19 knockout cell lines.
#CDK8 module = CDK8, CCNC, MED12, MED13
