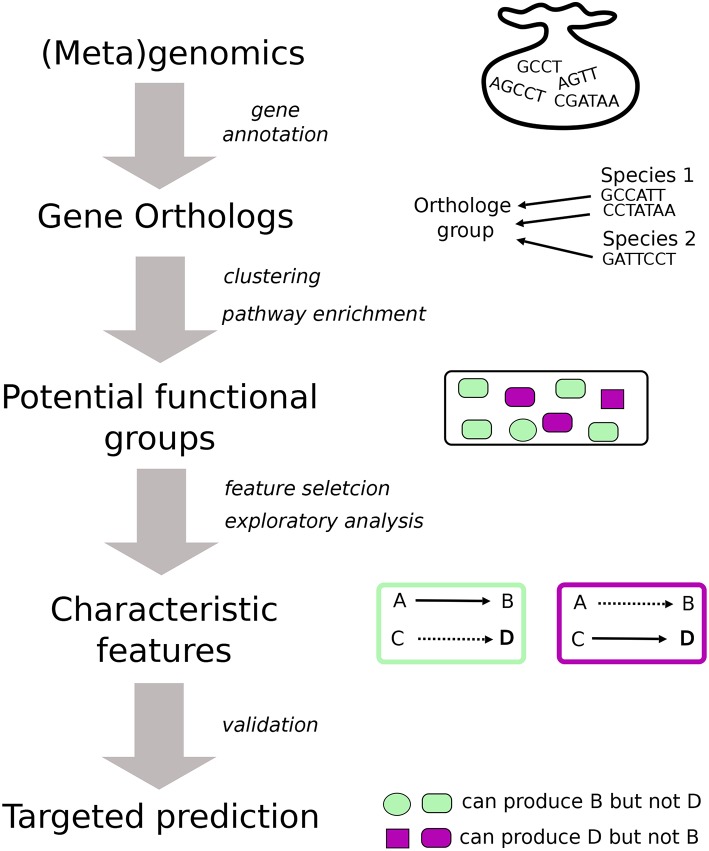
Figure 1.
Graphical representation of the general work-flow for the functional analysis. Starting point of the analysis is a gene annotation which is carried out using BlastKoala and GhostKoala in this study. We use the gene orthologs (KO's) to cluster species and samples based on their KO content. From the individual clusters we extract the characteristic features which leads to educated predictions about the functional potential of individual species and a community present in a sample. In the case of isolates, the predictions are confirmed using MetaDraft that does not rely on KO's.