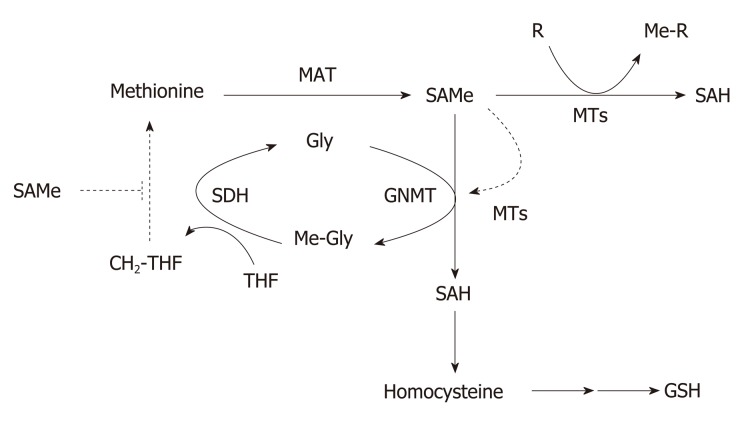
Figure 2.
Regulation of hepatic S-adenosylmethionine homeostasis. Hepatic S-adenosylmethionine (SAMe) content is regulated by the concerted activity of methionine adenosyltransferase (MAT) and glycine N-methyltransferase (GNMT). Methionine is mainly metabolized by the liver where is converted to SAMe by the enzyme MAT using ATP as co-substrate. SAMe, the main cellular methyl donor, is converted to S-adenosylhomocysteine (SAH) by a legion of methyltransferases (MTs) that catalyze the methylation of multiple substrates (DNA, proteins, phospholipids, small molecules, toxic and waist products). Excess SAMe is catabolized by GNMT, the most abundant hepatic MT, to prevent undesirable methylations. The GNMT-sarcosine dehydrogenase (SDH) pathway recycles the excess of methyl groups via generation of methylene-tetrahydrofolate (CH2-THF) and the methylation of homocysteine to regenerate methionine (not shown) to maintain SAMe homeostasis. SAH is converted to homocysteine, a metabolic crossroad that can be used for the regeneration of methionine (not shown) or the synthesis of glutathione depending on whether the concentration of SAMe is low or high, respectively. SAMe is an allosteric activator of GNMT and an inhibitor of the re-synthesis of methionine via the CH2-THF pathway (broken lines). CH2-THF: 5,10-methylene-tetrahydrofolate; Gly: Glycine; GNMT: Glycine N-methyltransferase; MAT: Methionine adenosyltransferase; Me-Gly: Methylglycine (sarcosine); Me-R: Methylated product; MTs: Methyltransferases; MTHF: 5-methyltetrahydrofolate; R: Methylation substrate; SAH: S-adenosylhomocysteine; SAMe: S-adenosylmethionine; SDH: Sarcosine dehydrogenase; THF: Tetrahydrofolate.