Abstract
Lung disease accounts for every sixth death globally. Profiling the molecular state of all lung cell types in health and disease is currently revolutionizing the identification of disease mechanisms and will aid the design of novel diagnostic and personalized therapeutic regimens. Recent progress in high-throughput techniques for single-cell genomic and transcriptomic analyses has opened up new possibilities to study individual cells within a tissue, classify these into cell types, and characterize variations in their molecular profiles as a function of genetics, environment, cell–cell interactions, developmental processes, aging, or disease. Integration of these cell state definitions with spatial information allows the in-depth molecular description of cellular neighborhoods and tissue microenvironments, including the tissue resident structural and immune cells, the tissue matrix, and the microbiome. The Human Cell Atlas consortium aims to characterize all cells in the healthy human body and has prioritized lung tissue as one of the flagship projects. Here, we present the rationale, the approach, and the expected impact of a Human Lung Cell Atlas.
Keywords: Human Cell Atlas, single-cell RNA sequencing, spatial transcriptomics, systems biology
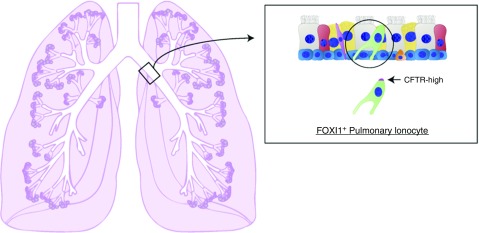
Lung disease is a leading cause of mortality in the United States and worldwide, with more than 7 million deaths attributed to lung disease annually (1). Although the lung has been reported to harbor at least 40 discrete cell types (2), the recent identification of the ionocyte as a novel epithelial cell type in human airway wall (Figure 1) shows that our knowledge of the cells in human lung is incomplete (3, 4). Therefore, further studies to systematically and comprehensively characterize all cell types in human lung tissue and the changes in cellular composition and function associated with initiation, progression, and resolution of human lung disease are urgently needed.
Figure 1.
The discovery of the pulmonary ionocyte. The first systematic analysis of the diversity of epithelial cells lining the airways by single-cell technology was recently reported in the mouse (3). In addition to finding an unexpected diversity of club, tuft, and goblet cells in the murine airway, a new airway epithelial cell type was discovered. This novel cell type specifically expresses the transcription factor Foxi1, and its composite gene expression profile resembles those of specialized ion-transporting cells in fish gills, frog skin, and mammalian kidney. As these diverse cell types are collectively referred to as ionocytes, the new pulmonary epithelial cell has been coined a “pulmonary ionocyte.” Surprisingly, these cells also expressed the majority of Cftr (cystic fibrosis transmembrane conductance regulator gene), whose mutation causes cystic fibrosis (CF). The investigators found that deletion of Foxi1 in mice resulted in the loss of mature ionocytes and significantly altered mucus viscosity. FOXI1+ pulmonary ionocytes were also found to line the airways of human lung and express high levels of CFTR. Their role in ion transport, regulation of mucus, and high CFTR expression suggests that pulmonary ionocytes play a critical role in CF biology and disease.
The cell is the fundamental unit of all living organisms (5). The cumulative function of all cells and their interactions with each other and with noncellular tissue components determines the physiology of any tissue or organ. Disease is believed to be a consequence of cell-intrinsic changes, changes in response to environmental insults, altered cell–cell communication, disbalance of cell type proportions, and/or perturbed architecture of tissues. Recent progress in single-cell analyses and accompanying computational methodologies provides an extraordinary opportunity to characterize individual cells and their spatial organization on the basis of their RNA or protein expression profile, allowing de novo classification of cell types and a comprehensive description of their dynamic phenotype. Combining these with rapidly evolving spatially resolving methods for the analysis of DNA, RNA, or protein profiles allows the interpretation or annotation of the cellular heterogeneity in a tissue context.
The Human Cell Atlas (HCA) Consortium is an international, collaborative effort aiming to define all human cell types in terms of their distinctive patterns of gene expression, physiological states, developmental trajectories, and spatial relationships in tissue (6). As part of this overarching effort, we intend to build a comprehensive atlas of human lung in a staged and integrated approach. Molecular profiles of single cells will be mapped computationally on a common coordinate framework of the entire organ in relation to spatial landmarks of both micro- and macro-anatomy, starting from the histological structure of cell neighborhoods, building up layers of increasingly larger units of spatial organization, up to the full organ.
The Lung Cell Atlas will reveal unprecedented insights about the identities, activities, and lineage relationships of all cells in healthy human lung, enabling the modeling of lung homeostatic circuitry (6). The Lung Cell Atlas will then serve as a reference point for the analysis of diseased lung tissue at single-cell resolution and will allow identification of the shifts in cellular repertoire, the changes in cellular states and phenotypes, and the altered cell–cell interactions that disrupt normal lung homeostasis and constitute disease. Comparing lung disease data with the lung development reference within the Human Lung Cell Atlas may reveal whether cellular changes in certain chronic lung diseases represent a failed recapitulation of organogenesis or the acquisition of entirely new pathologic programs governing cell fate and behavior. Thus, the generation of a high-resolution, comprehensive catalog of the changes in lung cellular composition and function in health and disease is expected to lead to development of novel cell- and disease-specific biomarkers and advancements in therapeutic strategies for lung disease.
Cellular and Anatomical Complexity of the Human Lung
The primary function of lung tissue—gas exchange between the body and its environment—is dependent on a specialized anatomy involving numerous distinct epithelial and endothelial cell populations, supported by specific tissue-resident leukocyte populations, and various mesenchymal cell types providing structural support. The unique anatomy of the lung is established during development by branching morphogenesis resulting in a tree of airways and alveoli, mirrored with trees of blood and lymphatic vessels. Air flows from the nasal cavity into the conducting airways, involving trachea and the main bronchi, which iteratively bifurcate into the branching bronchial tree. The human trachea and airways are composed of a pseudostratified epithelium on the luminal side, with a basal lamina lined with mesenchyme, cartilage, smooth muscle, blood vessels, immune cells, and other rare cell types (7). The airways end in terminal bronchioles, which then lead into composite respiratory units including the respiratory bronchioles, the alveolar duct, and the alveolar sac, where gas exchange occurs (7). The airways serve as a conduit for gases from the atmosphere to the alveoli but also have a role in filtering the inhaled air for solid particles and removing these in a process called mucociliary clearance, thereby protecting the distal saccular structures of the alveoli. Gas exchange happens in the alveolar unit, featuring ultrathin type-1 pneumocytes wrapped by a layer of capillaries, supported by interstitial cells such as matrix-producing fibroblasts and the pulmonary surfactant layer (8). Type-2 pneumocytes produce and secrete surfactant, maintain the fluid balance of the alveolar unit, and serve as local facultative progenitor cells for type-1 cells (9–11). Resident immune cells, particularly alveolar macrophages and peribronchial and perivascular “interstitial” macrophages, play important roles in orchestration of the immune response and in the maintenance of lung homeostasis (12, 13). Alveolar macrophages and alveolar epithelial cells express reciprocal sets of ligands and receptors, which prevent an excessive inflammatory response to inhaled particles. Several studies in mice have demonstrated that peribronchial macrophages consist of multiple ontologically and anatomically distinct subsets (14); however, little is known about their human counterparts.
Single-cell molecular studies profiled from a comprehensive sampling of human lungs will empower the mapping of all possible cellular identities (or states) onto a framework of a high-dimensional phenotypic cellular (gene and protein expression) landscape. Such a phenotypic landscape will describe cell type identity as stable intermediates within a continuum of possible states, including lineage relationships and potential transitory paths between different states. The power of this approach is elegantly shown in a recent review on the cellular and molecular pathways involved in lung organogenesis, where single-cell datasets from mouse lung development are used to identify and map cellular trajectories during the different stages of organogenesis (15). The exciting recent demonstration of genetic lineage tracing in human tissues using mitochondrial DNA mutations will now also enable the combination of lineage inference with gene expression or chromatin state profiles in human tissues (16). Thus, the use of mitochondrial DNA mutations as natural genetic barcodes may reveal the clonal architecture in human lung health and disease. The multiple snapshots of molecular identities present along the high-dimensional gene expression landscape can also result in the molecular description of unknown cellular states, which may have functions distinct from the traditional cell type definitions currently recognized. Similarly, we will likely encounter gradients of cellular phenotypes along spatial trajectories, as these “specialized” cellular states are driven by their microenvironment, as was recently reported for enterocytes in the mouse intestine (17).
All lung cells are contained within a regionally tailored and highly complex extracellular matrix that forms the basis of various distinct microenvironments, which is further defined by (the signals from) other cells in close proximity and potentially local variations in the microbiome. The effect of the microenvironment on the molecular phenotype of a cell has not been explored into great detail, although diseased human lung extracellular matrix, for instance, has been shown to drive a specific cellular transcriptome (18). Whether the identity of cells within the lung is determined by their specific location and function within the tissue and local microenvironment, by their developmental origin, or by a combination of both, is one of the key questions for the Human Lung Cell Atlas. Interestingly, it was recently reported that distinct developmental lineages can produce the same anatomical cell type by converging to a homogenous transcriptomic state in the final cell divisions of development, indicating that distinct developmental lineages can assume very similar molecular phenotypes as a function of their local niche (19). Location-dependent variation in the (molecular) state of cells in human lung has not been studied in detail yet, although in the mouse model, club cells have been shown to have a transcriptomic phenotype that is dependent on their position along the respiratory tract (3). The relative frequency of epithelial airway cell types varies along the respiratory tract (20). Submucosal glands are present from trachea to the small airways in human lung, but whether their composition is constant along the respiratory tract remains to be established. Similarly, it is unknown whether location within the lung affects the molecular phenotype of the cells that make up the alveolar unit. A unique feature of the lung is that it is a constantly moving organ, with dynamic changes in pressure and shape leading to presumably similar cells being exposed to very distinct mechanical cues—apical versus basal, central versus peripheral—and the cellular transcriptomic adaptations to these variables are completely unknown.
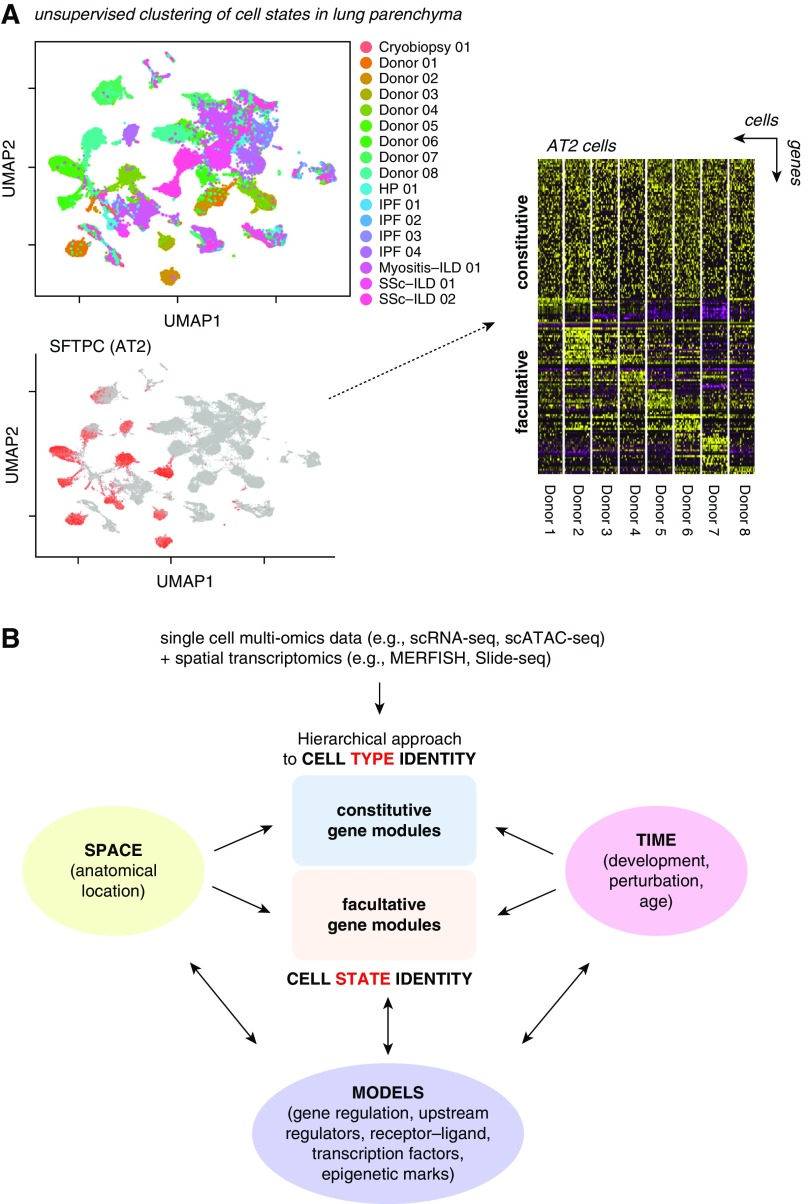
A complete atlas of the healthy human lung must sample the entire coordinate framework of the organ, both macro- and micro-anatomically, and capture the natural variation of cellular states within the healthy population (Figure 2). A comprehensive map of molecular cell states and the possible transitions between them will provide mechanistic insight into developmental processes and regenerative and repair capacity in the lung and their underlying regulatory mechanisms. The Human Lung Cell Atlas will map this landscape of molecular cell states onto a framework of spatial landmarks and enable the direct comparison of samples between multiple healthy and, eventually, diseased individuals.
Figure 2.
Cell types and cell states in the human lung cell atlas. (A) SFTPC-positive alveolar type-2 (AT2) cell clusters observed in lung parenchyma are shown in a dimension-reduced UMAP plot (left upper and lower panel). The heat map in the right panel shows constitutive genes defining AT2 cell type identity across eight healthy donors and clusters of genes that have been found to significantly differ between AT2 cells of different individuals. Hence, the constant features define the cell type, which can adopt multiple molecular phenotypes, or cell states, on the basis of the variable features. Adapted from Reference 53. (B) Data collected in the Human Lung Cell Atlas project will be used to define cell type identity in a hierarchical approach, which defines cell type by similarity metrics of their marker genes across many individuals. On the basis of these cell type identities and observed variation in facultative gene modules within the different cell types, we will generate predictive models of gene regulation that can be experimentally tested. These models will put (disease) genes in the context of their cell type–specific “gene regulatory environments” and predict the appearance of both constitutive and facultative gene modules as a function of time and space as occurring during lung development or regenerative and immune responses. HP = hypersensitivity pneumonitis; ILD = interstitial lung disease; IPF = idiopathic pulmonary fibrosis; MERFISH = multiplexed error-robust fluorescence in situ hybridization; scATAC-seq = single-cell Assay for Transposase-Accessible Chromatin sequencing; scRNA-seq = single-cell RNA sequencing; SSc = systemic sclerosis; UMAP = uniform manifold approximation and projection.
A Cellular Reference Map of Human Lung Development
The process of building the lung structure, and maturing the cells it contains, occurs over many weeks of embryonic development and beyond in humans and is believed to require numerous different cell–cell and cell–matrix interactions (15, 21). Many of these signaling and physical interactions occur between multiple progenitor cell types, which are specific to a developmental time window and do not exist in the adult organ. The epithelium is the best-characterized part of the developing lung, and at least three, apparently embryonic-specific, progenitor cell types have been detected. For example, the budding tip of the developing human lung epithelium was shown to be analogous to that of the mouse and to contain a major epithelial progenitor population required during branching morphogenesis (22, 23). However, there is no evidence that such cells exist in the mature lung. Characterization of the development-specific, or possibly developmentally enriched, progenitor cell states, and their (developmental) trajectories in the transcriptomic landscape, within the Human Cell Atlas Project will first and foremost provide insight into the basic mechanisms of lung development. In addition, it will provide specific and critical data allowing long-standing hypotheses regarding the recapitulation of developmental mechanisms in repair and disease states to be tested. Although the initial focus of the Human Lung Cell Atlas is on adult lung, it will integrate data from the Pediatric Cell Atlas (24) and initiatives focused on lung development, such as LungMAP or the recent National Institutes of Health (NIH)-funded DEVMAP.
Because of the progressive nature of branching morphogenesis, the developing lung epithelium contains a continuum of cells along the proximal–distal axis in progressive states of maturity from multipotent progenitor to fully mature mucociliary, or alveolar, epithelium. This is particularly significant given the low rates of cell turnover in the adult organ (25) and means that analysis of embryonic lung development will allow states of differentiation, particularly intermediate forms, to be analyzed transcriptionally in a way that will be very difficult in the adult lung. In addition, the developing human lung mesenchymal and leukocyte cell lineages are almost completely uncharacterized. For example, it is unknown when artery-venous fate arises in the lung endothelium, or when alveolar macrophages first become resident in the human lungs. A Developmental Lung Cell Atlas will provide baseline data describing the molecular phenotype of the cells in developing lung, facilitating their future functional characterization. It is important to note that the human lung continues to grow after birth well into puberty and beyond. Although the cellular changes during this period are completely unknown, they are considered to be of critical importance to development of lung disease later in life (26). The presence of bronchial-associated lymphoid tissue is rarely observed during fetal lung development and infrequently in newborns and infants in the absence of infection (27), has been reported to increase with age during the first few years of life (28, 29), and is absent in healthy adults after the age of 20 years (30) except in the context of specific inflammatory conditions (31). This illustrates the intimate interactions between a developing immune system, environmental challenges, and a maturing lung during early life, further emphasizing the need for inclusion of neonatal and pediatric studies in the Human Lung Cell Atlas.
The Human Lung Cell Atlas of Healthy Aging and Disease
The aging process manifests itself in age-related changes in body composition, the endocrine system, and vasculature, which are initially subclinical (32). The lung grows progressively until early adulthood, and lung function peaks between 18 and 25 years of age. After this peak, lung function declines, associated with structural remodeling of the lung, progressive loss of alveolar surface area, and enlargement of alveolar size (33, 34). In addition, mucociliary clearance is reduced with age in both upper and lower airways (35, 36). At the cellular level, hallmarks of aging include genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient-sensing mitochondrial dysfunction, stem-cell exhaustion, cellular senescence, and altered cell–cell communication (37), which is reflected in (bulk) gene expression patterns from lung (38). Cellular senescence contributes to alterations in extracellular matrix and is associated with physiological remodeling of the aging lung (39), and mitochondrial dysfunction has been observed in age-related lung disorders, including chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis (40). Integrating deep-tissue proteomics and single-cell transcriptomics, a first draft of an aging mouse lung atlas reported increased transcriptional noise, indicating loss of epigenetic control (41). Furthermore, age-associated changes in composition of the extracellular matrix, ciliated cell frequency, and a proinflammatory microenvironment were evident in both proteome and transcriptome (41).
To systematically and comprehensively analyze the effect of age on human lung cells, tissue sources used for the Human Lung Cell Atlas should therefore be collected over the entire adult age range from 20 to 80 years old. To distinguish natural variation from subclinical disease and to incorporate a history of relevant environmental and infectious exposures, we will need to obtain detailed clinical data, and potentially need to use biological characteristics of aging, such as telomere length (42) and the epigenetic clock (43), to calibrate the findings and understand natural variation and boundaries of health and disease. The promise of single-cell molecular profiling is to deliver unprecedented insight into the cellular origins of human disease. The human genome project and decades of genomic research have enabled investigations into the association of gene polymorphisms with disease. Using single-cell transcriptomics, we can now move from genetic and genomic associations to more mechanistic questions about disease and cell type–associated gene networks and reveal the cellular context of disease-associated polymorphisms (44).
The Approach of the Human Lung Cell Atlas: Toolbox and Resources
Single-Cell Transcriptomics
Single-cell RNA sequencing (scRNA-seq) aims to comprehensively capture and sequence the RNA contents of a single cell, allowing a description of transcriptional heterogeneity in cell populations. Stable transcriptional phenotypes can be contrasted with differentiation trajectories of cells by inferring an ordered sequence of transcriptional states in pseudotime. The key to the utility of scRNA-seq in tissue profiling has been the massive increase from 10s to more than 100,000 cells per experiment in less than a decade (45, 46), due to technological advances as recently reviewed (47). Comprehensive profiling of a tissue under specific (developmental, diseased) conditions requires a high throughput, capturing all cell types and states—including putative novel ones—at a threshold of sensitivity that is determined by both cell input and the observed variation in transcriptional profiles between discrete cell states. Rare cell types with a transcriptional phenotype that is not sufficiently discrete can be missed in unsupervised clustering, as exemplified by the neuroendocrine cells in the recent cell census of airway wall tissue (48). Although enrichment or purification of these cell types before scRNA-seq analysis can be applied, this does require prior knowledge of their identity. Atlasing studies therefore often combine a high-throughput approach for cell type and cell marker identification with in-depth profiling of purified subpopulations of special interest. To assess completeness of the single-cell sampling in silico, bulk profiles from scRNA-seq can be compared with true bulk RNA-seq data generated from adjacent regions of tissue. Another attractive alternative is the use of new high-density spatial transcriptomic tools, such as Slide-seq or High-Density Spatial Transcriptomics (HDST) (49, 50). These spatial tools can identify the microanatomical niches that are systematically undersampled and guide efforts for targeted enrichment.
Currently, sequencing of single cells from freshly dissociated tissue yields the greatest number of genes identified per cell. However, this comes at the cost of introducing potential dissociation biases, such as a stress response induced by enzymatic tissue digestion (51) or a bias in cell type composition, as cell types differ in their optimal conditions for release from the tissue (52, 53). An alternative technique is the isolation of nuclei from snap-frozen tissue, which are then assayed by single nuclear RNAseq protocols (snRNA-seq). In general, snRNA-seq can be performed on cryostat sections of lung tissue, with the advantage of improved tissue processing with less biased isolation, and enabling profiling of adjacent tissue or serial sections using targeted imaging techniques to put the snRNA-seq data into spatial context (49, 50, 54–57). Although single-nucleus RNA-seq detects fewer genes and unique transcripts than scRNA-seq, it allows the use of archived samples (58–61). On the whole, scRNA-seq and snRNA-seq data correlate well (62) and allow robust cell type identification as well as the derivation of developmental trajectories for the different cell types present in a tissue.
Beyond Single-Cell Transcriptomics
Although much of single-cell biology has focused on the transcriptome, epigenetic profiling of single cells can provide information on a cells’ past history and potential future states. Of the various epigenetic profiling methods available, Assay for Transposase-Accessible Chromatin (ATAC)-seq uses transposase tagging to identify regions of open chromatin and regulatory elements within the genome. Recently, single-cell ATAC-seq (scATAC-seq) has become available with the development of upfront bulk tagmentation, followed by DNA fragmentation at the single-cell level (63, 64). scATAC-seq has been shown to provide mechanistic insight into the regulatory mechanisms underlying cell states and their transitions (65). Differentiation trajectories of blood cells detected with scATAC-seq were similar to those from RNA but with earlier branching points, reflecting that the chromatin changes detected by ATAC accessibility precede expression at the RNA level (66). Thus, scATAC-seq and scRNA-seq will provide complementary insights into lung-cell states and the trajectories between them. As the cost of high-throughput scATAC protocols continues to reduce (67), we expect that this approach will contribute greatly to our understanding of lung development, cell type specification, and abnormality in disease.
Additional single-cell “omics” approaches, such as genome, methylome, proteome, and metabolome, are becoming available to complement transcriptomic measurements (68). The true size and shape of the cellular phenotypic landscape of any organ will only be revealed in a multiomic description of the cells and the noncellular components. Several multiomics single-cell approaches have been explored, including integrated single-cell measurement of chromatin accessibility; DNA methylation and transcriptome (69); integrated genetic, epigenetic, and transcriptomic measurement (70); and recently integrated scRNA-seq with genotype data to discover cell type–specific expression quantitative trait loci (71). Recent progress in single-cell profiling technologies has made it possible to measure protein and RNA levels within the same cell simultaneously (72–74), which enhances discovery of subtle cellular phenotypes, such as cell type subpopulations (75).
Imaging and Spatial Transcriptomics
Despite the advance of single-cell profiling, the function of individual cells within a tissue ultimately depends on their location within the tissue architecture and their interactions with the extracellular matrix and neighboring cells. Considering the morphological diversity of the lung, characterization of the exact location and local microenvironment where cells are obtained from is critically important. Techniques for measuring gene expression within lung tissue context range from spatial transcriptomics, microdissection approaches such as laser-capture microdissection, and in situ RNA hybridization, to in situ RNA sequencing. The main limitations on spatial mapping of cell states defined by high-dimensional gene expression patterns, however, are the number of genes detected on the same tissue sample and the shift from targeted to unbiased detection of gene expression. Spatial transcriptomic approaches measure gene expression in small voxels of tissue, with recent High-Density Spatial Transcriptomics (HDST) and Slide-seq technologies bringing the resolution down to around 1 to 10 μm (49, 50). A single-molecule fluorescence in situ hybridization (FISH)-based protocol to achieve high throughput is the multiplexed error-robust FISH (or MERFISH) technology (76), although application to the lung tissue might require further optimization (77). Another protocol suitable for multiplexed RNA species detection is the proximity ligation and in situ hybridization technology (78). Interestingly, proximity ligation and in situ hybridization is compatible with immunohistochemical applications and the use of antibodies on archived formalin-fixed paraffin embedded (FFPE) tissue. Similarly, detection of proteins and their post-translational modification with antibodies in combination with mRNA transcripts in tissue sections was also achieved using a mass cytometric approach (79). Finally, in situ RNA sequencing using padlock probes (77, 80) or fluorescence in situ RNA sequencing technology (81) is of interest. The recently developed STARmap method combined in situ amplification of mRNA transcripts with hydrogel tissue chemistry, allowing detection of around a thousand genes in millimeter scale tissue blocks after clearing of cells and lipids, while retaining relative location within the tissue blocks’ coordinate system (82). Integration of such methods with high-resolution transcriptomic single-cell profiling of matched tissue will likely be key to establish a true atlas of the cellular landscape of healthy or diseased human lung. A final technological challenge is to integrate proteomic data and transcriptomic data from single cells, which requires development of proteomic methods with single-cell sensitivity. Currently, a combination of antibody-labeling technologies or multiomics approaches, such as combining mass cytometry by time-of-flight (CyTOF) with spatially resolving FISH methodologies, remains the approach of choice.
Sources of Lung Tissue
Lung tissue for the Human Lung Cell Atlas is typically received from (routine) surgical resections, from biopsy extraction in routine clinical care, or in a research setting when involving healthy volunteers or lung transplantation programs. In this context, it is important to highlight the difficulty of obtaining normal lung parenchyma. The most common sources for lung parenchyma are either uninvolved edges of lung resections (mostly for cancer) or tissue obtained from deceased donors through organ procurement organizations, which all can be archived or processed fresh. Common sources of archived lung tissue are large tissue banks and repositories that have been established in many centers, mostly associated with academic, medical, or governmental organizations (83). Biobanks or core tissue facilities take care of patient information, obtain informed written consent for tissue donations and biopsy collections, and process and store tissue in an appropriate manner. They organize and monitor tissue distribution for scientific research, store relevant clinical information with respect to preserving patient anonymity, and provide centralized data storage. Specimens are collected according to site-specific protocols and legal regulations. To minimize tissue collection variability between different centers, common standard principles for tissue processing and collection are being developed. Live tissue from bronchoscopic biopsies, organ procurement organizations, or surgical resection needs special attention after collection. These specimens need to be transported immediately after acquisition to the research site for preparation of single-cell suspensions and downstream processing. Protocols for acquisition, transport, and processing of fresh tissue samples are harmonized between the HCA Lung working group members and are available through online platforms (such as https://www.protocols.io/groups/hca/publications).
Computational Challenges
The number of observations in a typical single-cell experiment is generally much larger than bulk RNA-seq data. Therefore, the underlying gene expression matrices, often encoded with genes as rows and cells as columns, pose novel and complex computational challenges because of their size. Moreover, so-called dropout events, in which an expressed gene is simply not detected because of lack of sensitivity, introduce high levels of sparsity into the data. Third, the increased resolution enables more precise capture of biological heterogeneity, which in turn leads to increased complexity in the data. Altogether, scalability, sparsity, and complexity in single-cell data pose major challenges when analyzing single-cell expression profiles.
Some popular scRNA-seq analysis frameworks, such as Scanpy (84), are designed to provide high scalability. Moreover, dimension reduction applications in molecular biology have evolved with the increased size and complexity of single-cell data. Linear dimension reduction by principal component analysis is commonly used for the visualization of bulk and initial scRNA-seq datasets. More recent approaches, using nonlinear two-dimensional embeddings, such as t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP), have helped visualize single-cell data and have recently been discussed with respect to quality as well as scalability (85, 86). These nonlinear approaches are particularly suitable for highly complex tissues, such as the lung, where a large number of different cell types exist (41, 53).
The increased number of cells profiled in a single experiment represents a computational challenge but also an opportunity for the application of more sophisticated modeling approaches, including machine learning (46). Deep learning, a subfield of machine learning, which has for instance revolutionized image analysis, is particularly powerful when large training datasets exist (87). Some of the recent, larger datasets now open up the possibility to apply deep-learning methods to scRNA-seq data. First, deep-learning algorithms have been developed and tailored toward scRNA-seq data with, for example, the goal of denoising scRNA-seq expression data (88). Machine learning approaches could be very useful for the detection of robust cell type–specific signatures and the integration of datasets generated on different platforms, where sequencing depth and transcript capture bias can vary dramatically depending on the protocol and experimental setup. As more data become available and datasets grow in size, we anticipate more machine learning applications for the analysis of scRNA-seq data.
Unbiased clustering approaches can be applied not only to scRNA-seq or scATAC-seq data but also to spatial techniques to uncover interactions between the cell types forming lung microniches, as was recently demonstrated for epithelial and mesenchymal cells (10, 78, 89). These tools will be particularly helpful for integration of high-throughput sequencing and spatial datasets. In combination with neural network–based approaches, we expect that additional data features such as morphometry—which can for example be used to robustly predict cell cycle stage (90)—will be integrated into spatial analysis to learn about combined cell state and communication.
Another promising computational approach is the statistical deconvolution of bulk RNA-seq data using scRNA-seq data to infer cell type proportions of a complex tissue sample (measured with bulk RNA-seq) by leveraging information from scRNA-seq data (91). It was recently shown in pancreatic datasets that cell-type heterogeneity represents a major covariate in bulk differential expression analysis (92). Lung tissue biopsies can show very high cell-type heterogeneity depending on the precise location of surgery (93). Therefore, publicly available lung bulk data from patients and control subjects is a particularly promising candidate for reanalysis using deconvolution techniques.
Opportunities to Build a Human Lung Cell Atlas
The opportunity to build a high-resolution single-cell and spatial atlas of the human body has generated enormous excitement in the scientific community. The global initiative to build the Human Cell Atlas, launched in 2016, now involves scientists from 62 countries, a number likely to increase. The HCA community is supported by many funding organizations and initiatives and organized into biological networks, including the Human Lung Cell Atlas. For example, in the specific area of the lung, the NIH has actively supported these efforts through programs, including LungMAP (94) in the healthy lung, and disease-focused programs, in particular the Cancer Moonshot Human Tumor Atlas Network (http://bit.ly/NCI-HumanTumorAtlas). Moreover, recognizing these emerging initiatives and the importance of supporting this ambitious new vision, the NIH Common Fund recently launched the Human BioMolecular Atlas Program (HuBMAP), which includes the lung among its organs of focus (https://commonfund.nih.gov/hubmap). Other relevant efforts are supported by the Medical Research Council (http://bit.ly/HCALungMRC) and Wellcome Trust (http://bit.ly/HCALungW) in the United Kingdom and by the Chan Zuckerberg Initiative (http://bit.ly/HCALungCZI1). The Human Lung Cell Atlas will be built by integration of the datasets provided by these and other initiatives. Working within the HCA thus ensures integration and international coordination across multiple funded efforts. Indeed, the HCA design is inherently inclusive and open, allowing parallel initiatives to contribute to the Human Cell Atlas, leveraging expertise, data, and technological developments between projects, organ systems, institutes, and countries. Consequently, the infrastructure that is under development within the HCA consortium will have a scope and reach beyond any of the organ-specific atlases, making the HCA well suited for a truly community-wide effort toward establishing a Human Cell Atlas.
Next Steps Toward Establishing a Human Lung Cell Atlas
The first cell censuses of upper airways and lung tissue have been published (48, 53, 95, 96), and more will follow with better coverage of regional differences and rare cell types, as evident from the progress meetings at the HCA meetings (http://bit.ly/HCALungTV1, http://bit.ly/HCALungTV2). These studies on single-cell profiling of healthy and diseased lung tissue mainly report scRNA-seq and ATAC-seq data (48, 53, 95, 96). With the anticipated increase in studies reporting molecular phenotypes of lung cells, the efforts of the Lung Biological Network within the HCA consortium is rapidly shifting from reporting on the progress of individual groups to coordinating shared and integrated analysis of the cumulative data within the HCA data coordination platform. The next breakthrough in establishing a Human Lung Cell Atlas will also come from a detailed spatial mapping of cell types onto the tissue architecture, the description of molecular cell states as a function of relative position in the lung along one of the many gradients present in the tissue or as a function of local cellular neighborhood, and the visualization of the cellular profiles in a spatial framework reflecting the structure of the lung. In addition, novel technological advances, especially in spatial mapping and computational approaches (e.g., toward multimodal data integration and combined visualization of profiling and spatial data) will remain an important topic for future meetings of the HCA consortium.
Joining the HCA Lung Biological Network
The HCA consortium is open for anyone in the respiratory community to join (https://www.humancellatlas.org/joinHCA), and open sharing of best practices and proven productive approaches to tissue acquisition, processing, data generation, and analysis as well as technological innovations will help bring the field forward. Progress will be presented at the HCA meetings but also at the annual meetings of the American Thoracic Society and the European Respiratory Society. Lung has been identified as one of the 12 priority organs and systems within the HCA initiative, with an initial focus on upper and lower airways as well as lung parenchyma (6). To achieve our ambition of establishing the Human Lung Cell Atlas, we call on widespread involvement of the respiratory community to contribute to this effort. This will allow the HCA Lung Biological Network to achieve the depth and breadth necessary to obtain a meaningful reference for basic, translational, and clinical respiratory scientists that may serve as an example for other less readily accessible organ systems within the HCA. To facilitate integration of datasets acquired by different research groups at geographically distinct sites, in multiple ethnicities, age groups, and sexes, we urgently need standardization in tissue procurement, processing, and storage, as well as in data generation, handling, storage, and analysis pipelines. For tissue acquisition, we have developed a series of standardized protocols specifically tested and validated for lung tissue and continue to expand these, making them available to the community through open access platforms such as protocols.io (https://www.protocols.io/groups/hca/publications). For processing and integration of data originating from multiple centers and divergent technological platforms, methodologies are being tested using the datasets from the initial lung scRNA-seq efforts. The computational approaches are also shared with the community, for instance through platforms such as Github (https://github.com/HumanCellAtlas). Methods for integration of genomic, transcriptomic, and proteomic datasets from the cellular branch of the Human Lung Cell Atlas with those acquired in the spatial branch will need to be further developed. For data storage and accessibility, the HCA lung project will tap into the HCA data coordination platform infrastructure that is currently in beta testing. One critical aspect for the Human Lung Cell Atlas to achieve its full impact will be the development of an interactive, tertiary HCA data portal (6) that allows exploration of the data by the respiratory and single-cell community. At this moment, interim data portals allow visualization and limited exploration of the currently available datasets (see for instance http://bit.ly/LungMap-scRNASeq [94], https://lungcellatlas.org [48], https://theislab.github.io/LungAgingAtlas [41], and https://www.nupulmonary.org/resources/ [53]). Incorporation of detailed demographic and clinical metadata from each tissue source in the HCA data coordination portal will be critical to allow the identification of individual and combined contributions of sex, ethnicity, age, respiratory health, and disease to the position of each cell type in the transcriptomic and proteomic landscape that defines the Human Lung Cell Atlas.
The Human Lung cell atlas holds the promise of becoming a resource of unsurpassed scale and detail, allowing the respiratory community to acquire novel insights into the biology of the lung, including—but not restricted to—its development, anatomy, and physiology in health and the changes thereof in lung disease.
Acknowledgments
Acknowledgment
The authors thank Dana Pe′er and Jennifer Rood for discussions on the paper outline and feedback on the manuscript.
Footnotes
Supported by the Helmholtz Association and the German Center for Lung Research (DZL) (H.B.S.); the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement 753039 (L.M.S.); U.K. Medical Research Council grant G0900424 (E.L.R.); National Institutes of Health (NIH) grants ES013995, HL071643, and AG049665, and Veterans Administration grant BX000201 and Department of Defense grant PR141319 (G.R.S.B.); NIH grants HL135124 and AI135964 and Department of Defense grant PR141319 (A.V.M.); NIH grants R01HL141852, R01HL127349, UHHL3123886, U01HL122626, and UG3TR002445, and Department of Defence grant PR151124 (N.K.); and the Netherlands Lung Foundation grants 5.1.14.020 and 4.1.18.226 (M.C.N.).
Author Contributions: Conception and outline: H.B.S., A.R., N.K., J.R., S.A.T., A.V.M., and M.C.N.; drafting the manuscript for important intellectual content: all authors.
Originally Published in Press as DOI: 10.1165/rcmb.2018-0416TR on April 17, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Gibson GJ, Loddenkemper R, Lundbäck B, Sibille Y. Respiratory health and disease in Europe: the new European Lung White Book. Eur Respir J. 2013;42:559–563. doi: 10.1183/09031936.00105513. [DOI] [PubMed] [Google Scholar]
- 2.Franks TJ, Colby TV, Travis WD, Tuder RM, Reynolds HY, Brody AR, et al. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc. 2008;5:763–766. doi: 10.1513/pats.200803-025HR. [DOI] [PubMed] [Google Scholar]
- 3.Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. Human Cell Atlas Meeting Participants. The Human Cell Atlas. eLife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Regev A, Teichmann S, Rozenblatt-Rosen O, Stubbington M, Ardlie K, Amit I, et al. The Human Cell Atlas White Paper. 2018 [accessed 2018 Nov 1]. Available from: http://arxiv.org/abs/1810.05192.
- 7.Hogan BLM, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CCW, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weibel ER. On the tricks alveolar epithelial cells play to make a good lung. Am J Respir Crit Care Med. 2015;191:504–513. doi: 10.1164/rccm.201409-1663OE. [DOI] [PubMed] [Google Scholar]
- 9.Whitsett JA, Wert SE, Weaver TE. Diseases of pulmonary surfactant homeostasis. Annu Rev Pathol. 2015;10:371–393. doi: 10.1146/annurev-pathol-012513-104644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, Desai TJ. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science. 2018;359:1118–1123. doi: 10.1126/science.aam6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature. 2018;555:251–255. doi: 10.1038/nature25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim HY, Lim SY, Tan CK, Thiam CH, Goh CC, Carbajo D, et al. Hyaluronan receptor LYVE-1-expressing macrophages maintain arterial tone through hyaluronan-mediated regulation of smooth muscle cell collagen Immunity 201849326–341.[Published erratum appears in Immunity 49:1191.] [DOI] [PubMed] [Google Scholar]
- 13.Gibbings SL, Thomas SM, Atif SM, McCubbrey AL, Desch AN, Danhorn T, et al. Three unique interstitial macrophages in the murine lung at steady state. Am J Respir Cell Mol Biol. 2017;57:66–76. doi: 10.1165/rcmb.2016-0361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan SYS, Krasnow MA. Developmental origin of lung macrophage diversity. Development. 2016;143:1318–1327. doi: 10.1242/dev.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitsett JA, Kalin TV, Xu Y, Kalinichenko VV. Building and regenerating the lung cell by cell. Physiol Rev. 2019;99:513–554. doi: 10.1152/physrev.00001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig LS, Lareau CA, Ulirsch JC, Christian E, Muus C, Li LH, et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell. 2019;176:1325–1339. doi: 10.1016/j.cell.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moor AE, Harnik Y, Ben-Moshe S, Massasa EE, Rozenberg M, Eilam R, et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell. 2018;175:1156–1167. doi: 10.1016/j.cell.2018.08.063. [DOI] [PubMed] [Google Scholar]
- 18.Parker MW, Rossi D, Peterson M, Smith K, Sikström K, White ES, et al. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Invest. 2014;124:1622–1635. doi: 10.1172/JCI71386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Packer JS, Zhu Q, Huynh C, Sivaramakrishnan P, Preston E. Dueck H, et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single cell resolution [preprint] bioRxiv. 2019 doi: 10.1126/science.aax1971. [accessed 2019 Mar 11]. Available from: https://www.biorxiv.org/content/10.1101/565549v2.abstract. [DOI] [PMC free article] [PubMed]
- 20.Tata PR, Rajagopal J. Plasticity in the lung: making and breaking cell identity. Development. 2017;144:755–766. doi: 10.1242/dev.143784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolić MZ, Sun D, Rawlins EL. Human lung development: recent progress and new challenges. Development. 2018;145:163485. doi: 10.1242/dev.163485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikolić MZ, Johnson J-A, Sun D, Caritg O, Laresgoiti U, Brady J, et al. Development of a genetically modifiable epithelial in-vitro culture system from human embryonic lung epithelial stem cells: towards human lung regeneration in end-stage respiratory failure. Lancet. 2017;389:S74. [Google Scholar]
- 23.Miller AJ, Hill DR, Nagy MS, Aoki Y, Dye BR, Chin AM, et al. In vitro induction and in vivo engraftment of lung bud tip progenitor cells derived from human pluripotent stem cells. Stem Cell Reports. 2018;10:101–119. doi: 10.1016/j.stemcr.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor DM, Aronow BJ, Tan K, Bernt K, Salomonis N, Greene CS, et al. The pediatric cell atlas: defining the growth phase of human development at single-cell resolution. Dev Cell. 2019;49:10–29. doi: 10.1016/j.devcel.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teixeira VH, Nadarajan P, Graham TA, Pipinikas CP, Brown JM, Falzon M, et al. Stochastic homeostasis in human airway epithelium is achieved by neutral competition of basal cell progenitors. eLife. 2013;2:e00966. doi: 10.7554/eLife.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bush A. Lung development and aging. Ann Am Thorac Soc. 2016;13:S438–S446. doi: 10.1513/AnnalsATS.201602-112AW. [DOI] [PubMed] [Google Scholar]
- 27.Gould SJ, Isaacson PG. Bronchus-associated lymphoid tissue (BALT) in human fetal and infant lung. J Pathol. 1993;169:229–234. doi: 10.1002/path.1711690209. [DOI] [PubMed] [Google Scholar]
- 28.Tschernig T, Kleemann WJ, Pabst R. Bronchus-associated lymphoid tissue (BALT) in the lungs of children who had died from sudden infant death syndrome and other causes. Thorax. 1995;50:658–660. doi: 10.1136/thx.50.6.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heier I, Malmström K, Sajantila A, Lohi J, Mäkelä M, Jahnsen FL. Characterisation of bronchus-associated lymphoid tissue and antigen-presenting cells in central airway mucosa of children. Thorax. 2011;66:151–156. doi: 10.1136/thx.2010.149591. [DOI] [PubMed] [Google Scholar]
- 30.Hiller AS, Tschernig T, Kleemann WJ, Pabst R. Bronchus-associated lymphoid tissue (BALT) and larynx-associated lymphoid tissue (LALT) are found at different frequencies in children, adolescents and adults. Scand J Immunol. 1998;47:159–162. doi: 10.1046/j.1365-3083.1998.00276.x. [DOI] [PubMed] [Google Scholar]
- 31.Tschernig T, Pabst R. Bronchus-associated lymphoid tissue (BALT) is not present in the normal adult lung but in different diseases. Pathobiology. 2000;68:1–8. doi: 10.1159/000028109. [DOI] [PubMed] [Google Scholar]
- 32.Partridge L, Deelen J, Slagboom PE. Facing up to the global challenges of ageing. Nature. 2018;561:45–56. doi: 10.1038/s41586-018-0457-8. [DOI] [PubMed] [Google Scholar]
- 33.Ochs M, Nyengaard JR, Jung A, Knudsen L, Voigt M, Wahlers T, et al. The number of alveoli in the human lung. Am J Respir Crit Care Med. 2004;169:120–124. doi: 10.1164/rccm.200308-1107OC. [DOI] [PubMed] [Google Scholar]
- 34.Hecker L. Mechanisms and consequences of oxidative stress in lung disease: therapeutic implications for an aging populace. Am J Physiol Lung Cell Mol Physiol. 2018;314:L642–L653. doi: 10.1152/ajplung.00275.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Proença de Oliveira-Maul J, Barbosa de Carvalho H, Goto DM, Maia RM, Fló C, Barnabé V, et al. Aging, diabetes, and hypertension are associated with decreased nasal mucociliary clearance. Chest. 2013;143:1091–1097. doi: 10.1378/chest.12-1183. [DOI] [PubMed] [Google Scholar]
- 36.Svartengren M, Falk R, Philipson K. Long-term clearance from small airways decreases with age. Eur Respir J. 2005;26:609–615. doi: 10.1183/09031936.05.00002105. [DOI] [PubMed] [Google Scholar]
- 37.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Vries M, Faiz A, Woldhuis RR, Postma DS, de Jong TV, Sin DD, et al. Lung tissue gene-expression signature for the ageing lung in COPD. Thorax. doi: 10.1136/thoraxjnl-2017-210074. [online ahead of print]; 6 Dec 2017 DOI: 10.1136/thoraxjnl-2017-210074. [DOI] [PubMed] [Google Scholar]
- 39.Calhoun C, Shivshankar P, Saker M, Sloane LB, Livi CB, Sharp ZD, et al. Senescent cells contribute to the physiological remodeling of aged lungs. J Gerontol A Biol Sci Med Sci. 2016;71:153–160. doi: 10.1093/gerona/glu241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meiners S, Eickelberg O, Königshoff M. Hallmarks of the ageing lung. Eur Respir J. 2015;45:807–827. doi: 10.1183/09031936.00186914. [DOI] [PubMed] [Google Scholar]
- 41.Angelidis I, Simon LM, Fernandez IE, Strunz M, Mayr CH, Greiffo FR, et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun. 2019;10:963. doi: 10.1038/s41467-019-08831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Everaerts S, Lammertyn EJ, Martens DS, De Sadeleer LJ, Maes K, van Batenburg AA, et al. The aging lung: tissue telomere shortening in health and disease. Respir Res. 2018;19:95. doi: 10.1186/s12931-018-0794-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19:371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 44.Hemberg M. Summing up the parts of the hypothalamus. Nat Neurosci. 2017;20:378–379. doi: 10.1038/nn.4515. [DOI] [PubMed] [Google Scholar]
- 45.Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13:599–604. doi: 10.1038/nprot.2017.149. [DOI] [PubMed] [Google Scholar]
- 46.Angerer P, Simon L, Tritschler S, Alexander Wolf F, Fischer D, Theis FJ. Single cells make big data: new challenges and opportunities in transcriptomics. Curr Opin Syst Biol. 2017;4:85–91. [Google Scholar]
- 47.Chen X, Teichmann SA, Meyer KB. From tissues to cell types and back: single-cell gene expression analysis of tissue architecture. Annu Rev Biomed Data Sci. 2018;1:29–51. [Google Scholar]
- 48.Vieira Braga FA, Kar G, Berg M, Carpaij OA, Polanski K, Simon LM, et al. A cellular census of healthy lung and asthmatic airway wall identifies novel cell states in health and disease [preprint] bioRxiv. 2019 doi: 10.1038/s41591-019-0468-5. [accessed 2019 Mar 11]. Available from: https://www.biorxiv.org/content/10.1101/527408v1.abstract. [DOI] [PubMed] [Google Scholar]
- 49.Vickovic S, Eraslan G, Salmén F.Klughammer J, Stenbeck L, Äijö T, et al. High-density spatial transcriptomics arrays for in situ tissue profiling [preprint] bioRxiv 2019[accessed 2019 Mar 11]. Available from: https://www.biorxiv.org/content/10.1101/563338v1.abstract [DOI] [PMC free article] [PubMed]
- 50.Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van den Brink SC, Sage F, Vértesy Á, Spanjaard B, Peterson-Maduro J, Baron CS, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- 52.Cohen M, Giladi A, Gorki A-D, Solodkin DG, Zada M, Hladik A, et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell. 2018;175:1031–1044. doi: 10.1016/j.cell.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. doi: 10.1164/rccm.201712-2410OC. 10.1164/rccm.201712-2410OC [online ahead of print] 15 Dec 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 55.Pinskiy V, Jones J, Tolpygo AS, Franciotti N, Weber K, Mitra PP. High-throughput method of whole-brain sectioning, using the tape-transfer technique. PLoS One. 2015;10:e0102363. doi: 10.1371/journal.pone.0102363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carow B, Hauling T, Qian X, Kramnik I, Nilsson M, Rottenberg ME. Spatial and temporal localization of immune transcripts defines hallmarks and diversity in the tuberculosis granuloma [preprint] bioRxiv. 2019 doi: 10.1038/s41467-019-09816-4. [accessed 2019 Mar 11]. Available from: https://www.biorxiv.org/content/10.1101/509547v1.abstract. [DOI] [PMC free article] [PubMed]
- 57.Ziegenhain C, Vieth B, Parekh S, Hellmann I, Enard W. Quantitative single-cell transcriptomics. Brief Funct Genomics. 2018;17:220–232. doi: 10.1093/bfgp/ely009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lacar B, Linker SB, Jaeger BN, Krishnaswami SR, Barron JJ, Kelder MJE, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Habib N, Li Y, Heidenreich M, Swiech L, Avraham-Davidi I, Trombetta JJ, et al. Div-seq: single-nucleus RNA-seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krishnaswami SR, Grindberg RV, Novotny M, Venepally P, Lacar B, Bhutani K, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc. 2016;11:499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lake BB, Codeluppi S, Yung YC, Gao D, Chun J, Kharchenko PV, et al. A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Sci Rep. 2017;7:6031. doi: 10.1038/s41598-017-04426-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, Miragaia RJ, Natarajan KN, Teichmann SA. A rapid and robust method for single cell chromatin accessibility profiling. Nat Commun. 2018;9:5345. doi: 10.1038/s41467-018-07771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen X, Litzenburger U, Wei Y, Schep AN, LaGory EL, Choudhry H, et al. Joint single-cell DNA accessibility and protein epitope profiling reveals environmental regulation of epigenomic heterogeneity. Nat Commun. 2018;9:4590. doi: 10.1038/s41467-018-07115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, et al. Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell. 2018;173:1535–1548. doi: 10.1016/j.cell.2018.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mezger A, Klemm S, Mann I, Brower K, Mir A, Bostick M, et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat Commun. 2018;9:3647. doi: 10.1038/s41467-018-05887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Colomé-Tatché M, Theis FJ. Statistical single cell multi-omics integration. Curr Opin Syst Biol. 2018;7:54–59. [Google Scholar]
- 69.Clark SJ, Argelaguet R, Kapourani C-A, Stubbs TM, Lee HJ, Alda-Catalinas C, et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Commun. 2018;9:781. doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van der Wijst MGP, Brugge H, de Vries DH, Deelen P, Swertz MA, Franke L LifeLines Cohort Study; BIOS Consortium. Single-cell RNA sequencing identifies celltype-specific cis-eQTLs and co-expression QTLs. Nat Genet. 2018;50:493–497. doi: 10.1038/s41588-018-0089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valdes AM, Glass D, Spector TD. Omics technologies and the study of human ageing. Nat Rev Genet. 2013;14:601–607. doi: 10.1038/nrg3553. [DOI] [PubMed] [Google Scholar]
- 73.Darmanis S, Gallant CJ, Marinescu VD, Niklasson M, Segerman A, Flamourakis G, et al. Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell Rep. 2016;14:380–389. doi: 10.1016/j.celrep.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35:936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 75.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moffitt JR, Hao J, Wang G, Chen KH, Babcock HP, Zhuang X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci USA. 2016;113:11046–11051. doi: 10.1073/pnas.1612826113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moor AE, Itzkovitz S. Spatial transcriptomics: paving the way for tissue-level systems biology. Curr Opin Biotechnol. 2017;46:126–133. doi: 10.1016/j.copbio.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 78.Nagendran M, Riordan DP, Harbury PB, Desai TJ. Automated cell-type classification in intact tissues by single-cell molecular profiling. eLife. 2018;7:e30510. doi: 10.7554/eLife.30510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schulz D, Zanotelli VRT, Fischer JR, Schapiro D, Engler S, Lun X-K, et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst. 2018;6:531. doi: 10.1016/j.cels.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ke R, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 81.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10:442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 2018;361:eaat5691. doi: 10.1126/science.aat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haga SB, Beskow LM. Ethical, legal, and social implications of biobanks for genetics research. Adv Genet. 2008;60:505–544. doi: 10.1016/S0065-2660(07)00418-X. [DOI] [PubMed] [Google Scholar]
- 84.Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. doi: 10.1038/nbt.4314. 10.1038/nbt.4314 [online ahead of print] 3 Dec 2018. [DOI] [PubMed] [Google Scholar]
- 86.Kobak D, Berens P. The art of using t-SNE for single-cell transcriptomics [preprint] bioRxiv. 2018. [accessed 2019 Mar 11]. Available from: https://www.biorxiv.org/content/10.1101/453449v1.abstract. [DOI] [PMC free article] [PubMed]
- 87.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 88.Eraslan G, Simon LM, Mircea M, Mueller NS, Theis FJ. Single-cell RNA-seq denoising using a deep count autoencoder. Nat Commun. 2019;10:390. doi: 10.1038/s41467-018-07931-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 2017;170:1134–1148. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eulenberg P, Köhler N, Blasi T, Filby A, Carpenter AE, Rees P, et al. Reconstructing cell cycle and disease progression using deep learning. Nat Commun. 2017;8:463. doi: 10.1038/s41467-017-00623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol. 2013;25:571–578. doi: 10.1016/j.coi.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baron M, Veres A, Wolock SL, Faust AL, Gaujoux R, Vetere A, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baharom F, Thomas S, Rankin G, Lepzien R, Pourazar J, Behndig AF, et al. Dendritic cells and monocytes with distinct inflammatory responses reside in lung mucosa of healthy humans. J Immunol. 2016;196:4498–4509. doi: 10.4049/jimmunol.1600071. [DOI] [PubMed] [Google Scholar]
- 94.Ardini-Poleske ME, Clark RF, Ansong C, Carson JP, Corley RA, Deutsch GH, et al. LungMAP Consortium. LungMAP: the molecular atlas of lung development program. Am J Physiol Lung Cell Mol Physiol. 2017;313:L733–L740. doi: 10.1152/ajplung.00139.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ordovas-Montanes J, Dwyer DF, Nyquist SK, Buchheit KM, Vukovic M, Deb C, et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature. 2018;560:649–654. doi: 10.1038/s41586-018-0449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, et al. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight. 2016;1:e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]