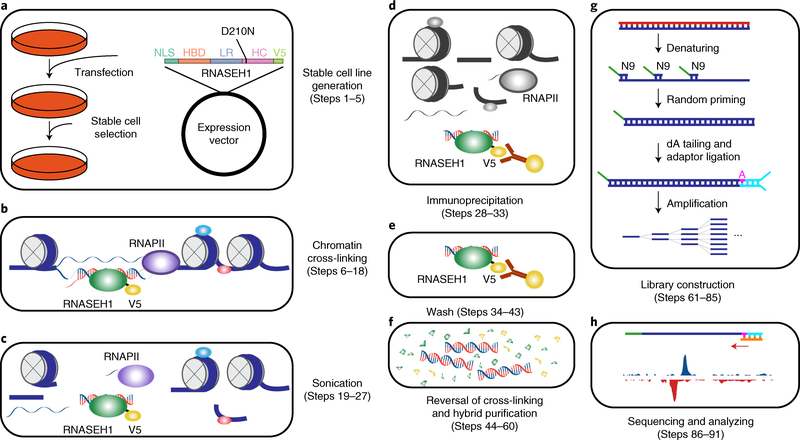
Fig. 1 |. Overview of the R-ChIP workflow.
a, Cells are transfected with an expression vector containing a mutant RNASEH1 (D210N) gene with a V5 tag at its N terminus, followed by drug selection to obtain a stable cell line expressing the mutant protein. b, Cells from a are first cross-linked to stabilize DNA–protein interactions. RNA and DNA molecules are colored in red and blue, respectively. Proteins are shown as ellipses. c, Nuclei are isolated and sheared by sonication. d, RNASEH1 mutant associated with RNA–DNA hybrids is immunoprecipitated with V5 antibody-conjugated beads. e, Beads are washed to remove nonspecifically bound chromatin fragments. f, RNA–DNA hybrids are recovered by reversal of cross-linking and purification. g, RNA–DNA hybrids are denatured, and the resultant ssDNA is converted into dsDNA by extension with a primer containing a 9-nt random sequence at the 3′ end. dA is added to the 3′ end of the extension product, followed by adaptor ligation (shown in cyan) to only one end, as the other end is blocked by the 5′ overhang generated by the N9 primer. PCR amplification is then performed for library construction. h, Single-end sequencing is performed and the results are analyzed. dA, deoxyadenosine triphosphate; HBD, RNA–DNA hybrid binding domain; HC, RNA–DNA hybrid catalytic domain; LR, linker region; NLS, nuclear localization signal; RNAPII, RNA polymerase II; ssDNA, single-stranded DNA; V5, V5 tag.