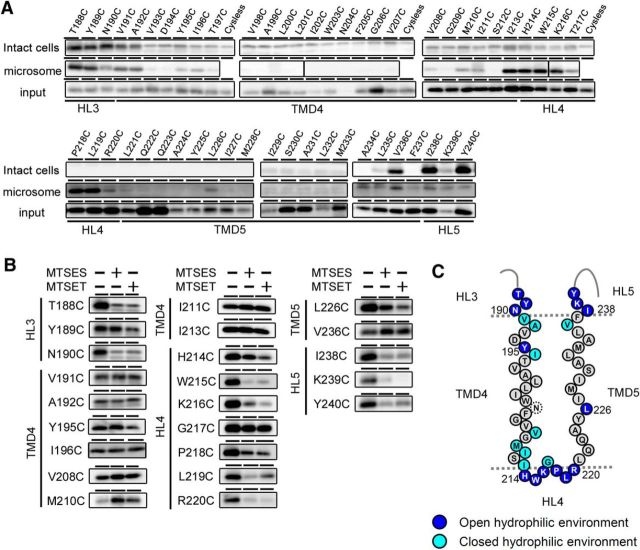
Figure 3.
SCAM analysis of single-Cys mt PS1s mutated around TMD4 and TMD5. A, Biotinylation experiment using MTSEA-biotin in intact cells (top panels) and microsomes (middle panels). Amounts of PS1 NTF in the input fraction are shown in the bottom panels. Putative domains are indicated below the panels. B, Labeling competition of single-Cys mt PS1s that were labeled by MTSEA-biotin in A was performed after preincubation with negatively charged MTSES or positively charged MTSET. Putative domains are indicated at the left of the panels. C, Summary of the biotinylation experiments using MTSEA-biotin and competition experiments using charged MTS reagents. All charged reagents were accessible to the Cys-substituted amino acids shown as dark blue circles (open hydrophilic environment). Residues in which labeling were not competed by MTSES or MTSET are shown as light blue circles (closed hydrophilic environment). Residues that were not labeled by MTSEA-biotin are shown as black letters in gray circles. N204C (dotted circle) was not endoproteolyzed.