Figure 5.
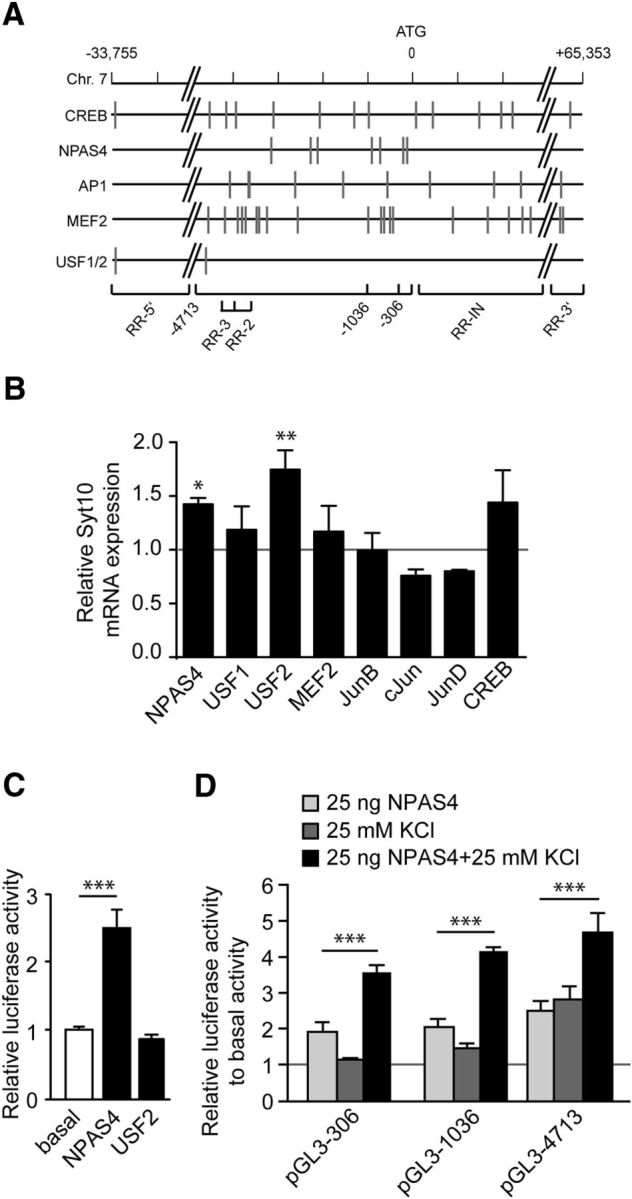
The Syt10 gene harbors functional binding sites for NPAS4. A, Graphic representation of the rat Syt10 gene with predicted transcription factor binding sites (indicated by gray vertical lines) of factors known to be involved in differential expression after synaptic activity (CREB1, NPAS4, AP1, MEF2, USF1/2). B, Quantitative RT-PCR for Syt10 mRNA expression levels from RH neurons transfected at 5 DIV with an overexpression construct for NPAS4, USF1, USF2, MEF2, JunB, cJun, JunD, or CREB. Cells were lysed 24 h (6 DIV) after transfection. All values were normalized to the respective control condition (transfection with an empty vector; gray line). Only for NPAS4 and USF2, a significant upregulation of Syt10 was observed. Statistical analysis was performed using a one-way ANOVA followed by Tukey post hoc test. *p ≤ 0.05, **p ≤ 0.01; N = 2, n = 3. C, Luciferase activity of RH neurons transfected at 5 DIV with an empty vector, 25 ng NPAS4 or 25 ng USF2 and lysed 48 h later. Firefly relative units are normalized to the basal values (mock transfected cells). NPAS4 increased the luciferase activity of the Syt10 promoter fragment −4713, whereas USF2 had no effect. Statistical analysis was performed using a one-way ANOVA followed by Tukey post hoc test. ***p ≤ 0.001; N = 3, n = 3. D, Luciferase activity of RH neurons that had been transfected with the indicated Syt10 promoter fragments and with 25 ng NPAS4 followed by KCl stimulation. Statistical analysis was performed using a one-way ANOVA followed by Tukey post hoc test. ***p ≤ 0.001; N = 2, n = 3.
