Figure 1.
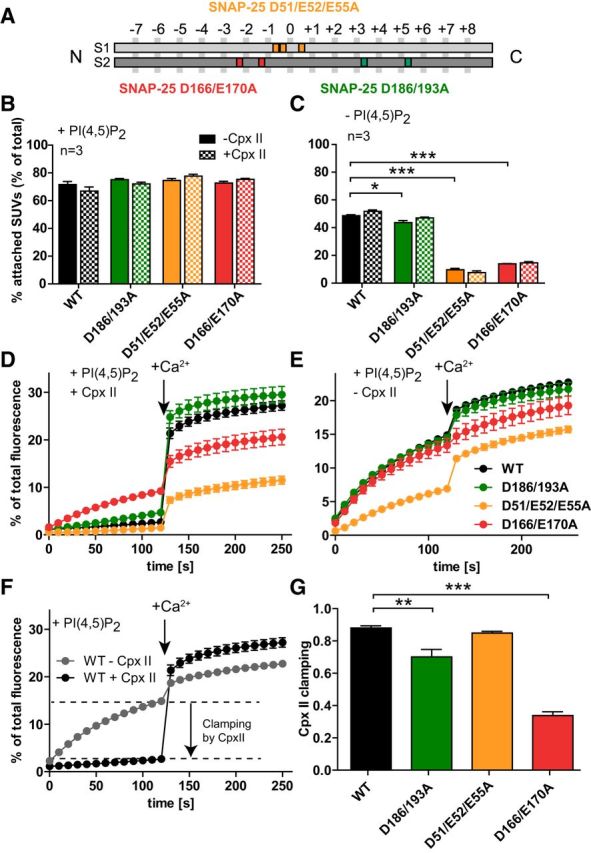
Vesicle–vesicle attachment and fusion in vitro are inhibited by the SNAP-25B mutations D166/E170A and D51/E52/E55A. A, Schematic representation of the mutants SNAP-25B D51/E52/E55A (orange), which is located around the hydrophilic zero layer of the SNARE helix 1 (S1), SNAP-25B D166A/E170A (red), and SNAP-25B D186/D193A (green), both located on the SNARE helix 2 (S2). B, C, t-SNARE-GUVs were mixed with 3H-labeled v-SNARE/syt-1-SUVs with and without Cpx II in the presence (B) or absence (C) of PI(4,5)P2. Samples were incubated for 5 min on ice to allow SUV:GUV attachment, followed by centrifugation to reisolate GUV. 3H-labeled SUVs that were attached to GUVs were quantified and normalized to total input. D, E, t-SNARE-GUVs containing SNAP-25 WT (black), SNAP-25B D186/D193A (green), SNAP-25B D51/E52/E55A (orange), or SNAP-25B D166/E170A (red) were mixed with v-SNARE-SUVs in the presence (D) or absence (E) of Cpx II. The increase of Atto488 fluorescence due to lipid mixing was monitored. After 2 min at 37°C, Ca2+ was added to a final concentration of 100 μm and the measurement continued for another 2 min. Values were normalized to the maximum fluorescence signal after detergent lysis. F, Direct comparison of lipid mixing for WT SNAP-25 in the absence (gray) and in the presence (black) of Cpx II. Cpx II clamps most of the prestimulation membrane fusion. G, Fractional clamping effect of Cpx II for the different constructs. Note that D166/E170A inhibits Cpx II clamping. All graphs display mean ± SEM (n = 3). *p < 0.05, **p < 0.01; ***p < 0.001 (one-way ANOVA, significance was calculated using Dunnett's post test).
