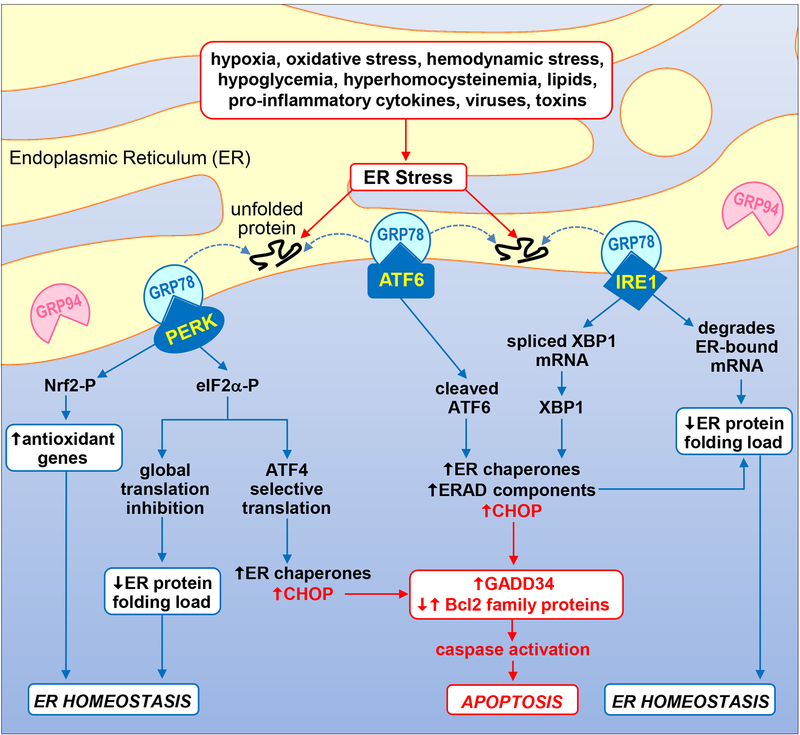
1.
Schematic diagram illustrating the UPR signal transduction pathway. Numerous pathophysiological stimuli result in the accumulation of unfolded and misfolded proteins in the ER lumen causing ER stress and the activation of the UPR. The presence of improperly folded proteins leads to the dissociation of GRP78 from the UPR sensors PERK, ATF6, and IRE1, and their activation. Subsequently, the kinase PERK phosphorylates eIF2α and Nrf2 to repress global protein synthesis and oxidative stress, respectively. Interestingly, phosphorylated eIF2α favors the cap-independent translation of the transcription factor ATF4 which promotes the expression of ER chaperones such as GRP78 and GRP94. Upon activation, the transcription factor ATF6 induces the expression of genes involved in protein folding activity (ER chaperones) and protein degradation (ERAD components). In addition, IRE1 splices XBP1 mRNA to mature XBP1 mRNA which is then translated to a potent transcription factor that stimulates the expression of ER chaperones and enzymes that target misfolded proteins for ERAD. IRE1 also degrades certain ER-localized mRNAs through regulated IRE1-dependent decay. Collectively, these actions function in an adaptive manner to reduce the ER protein folding load and redox stress in order to restore ER homeostasis. However, if ER stress is too severe or protracted, all three UPR sensors can induce the expression of the transcription factor CHOP which promotes caspase activation and apoptosis by increasing the expression of pro-apoptotic GADD34 and pro-apoptotic Bcl2 family members while suppressing the expression of anti-apoptotic Bcl2 proteins.