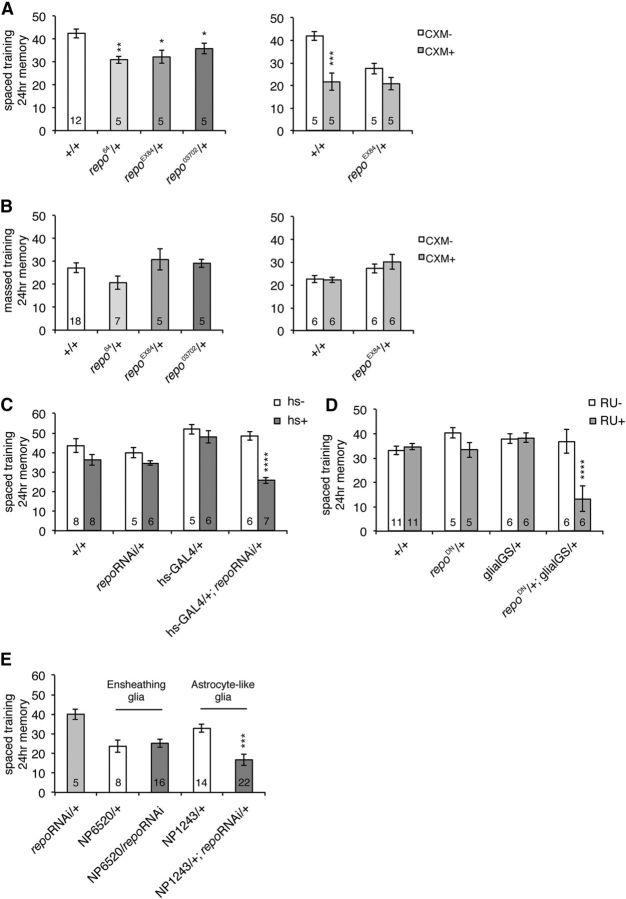
Figure 3.
Repo is required for LTM formation. A, Left, repo hypomorphic mutants are defective for memory 24 h after spaced training. Experiments were performed comparing single mutants to wild-type. *p < 0.05; **p < 0.01 compared with +/+ by t test. Right, CXM treatment does not reduce 24 h memory in repo mutants. Two-way ANOVA indicates significant differences due to genotype (F(1,16) = 7.448, p = 0.0149), CXM feeding (F(1,16) = 22.90, p = 0.0002), and interaction between genotype and CXM feeding (F(1,16) = 5.639, p = 0.0304). Bonferroni post hoc tests reveal significant differences due to CXM feeding in wild-type (***p < 0.001), but not repoEX84 mutants. B, Left, 24 h memory formed after massed training. Right, Effects of CXM feeding on 24 h memory after massed training. C, Acute knockdown of repo disrupts LTM. A 37°C heat shock (hs+) for 30 min significantly disrupted 1 d memory after spaced training in hs-GAL4/+; UAS-repoRNAi/+ flies. Two-way ANOVA demonstrates significant differences due to genotype (F(3,43) = 9.189, p < 0.0001), heat-shock regimen (F(1,43) = 26.90, p < 0.0001), and interaction between genotype and heat shock (F(3,43) = 5.245, p = 0.0036). ****p < 0.0001 compared with hs−, determined by Bonferroni post hoc tests. D, Acute overexpression of dominant-negative repo in glia disrupts LTM. The glial-GS driver was used to overexpress dominant-negative repo in the presence of RU486 in a UAS-repoΔAD302/+; glial-GS-GAL4/+ line. RU486 (1 mm) was fed to flies from 3 d before training until testing. Two-way ANOVA indicates significant differences due to genotype (F(3,48) = 7.347, p = 0.0004), RU feeding (F(1,48) = 11.81, p = 0.0012), and interaction between genotype and feeding (F(3,48) = 8.020, p = 0.0002). ****p < 0.0001 compared with RU−, as determined by Bonferroni post hoc tests. E, repo knockdown in astrocyte-like glia (NP1243/+; repo RNAi/+) disrupts LTM, while knockdown in ensheathing glia (NP6520/repoRNAi) does not. One-way ANOVA indicates significant differences due to genotype (F(4,60) = 7.979, p < 0.0001). Tukey's multiple-comparison tests reveal significant differences between NP1243/+; repo RNAi/+ and both repoRNAi/+ and NP1243/+ controls (***p < 0.001), but no significant differences between NP6520/repoRNAi and the NP6520/+ control.