Abstract
The proper growth and arborization of dendrites in response to sensory experience are essential for neural connectivity and information processing in the brain. Although neuronal activity is important for sculpting dendrite morphology, the underlying molecular mechanisms are not well understood. Here, we report that cyclin-dependent kinase 5 (Cdk5)-mediated transcriptional regulation is a key mechanism that controls activity-dependent dendrite development in cultured rat neurons. During membrane depolarization, Cdk5 accumulates in the nucleus to regulate the expression of a subset of genes, including that of the neurotrophin brain-derived neurotrophic factor, for subsequent dendritic growth. Furthermore, Cdk5 function is mediated through the phosphorylation of methyl-CpG-binding protein 2, a key transcriptional repressor that is mutated in the mental disorder Rett syndrome. These findings collectively suggest that the nuclear import of Cdk5 is crucial for activity-dependent dendrite development by regulating neuronal gene transcription during neural development.
SIGNIFICANCE STATEMENT Neural activity directs dendrite development through the regulation of gene transcription. However, how molecular signals link extracellular stimuli to the transcriptional program in the nucleus remains unclear. Here, we demonstrate that neuronal activity stimulates the translocation of the kinase Cdk5 from the cytoplasmic compartment into the nucleus; furthermore, the nuclear localization of Cdk5 is required for dendrite development in cultured neurons. Genome-wide transcriptome analysis shows that Cdk5 deficiency specifically disrupts activity-dependent gene transcription of bdnf. The action of Cdk5 is mediated through the modulation of the transcriptional repressor methyl-CpG-binding protein 2. Therefore, this study elucidates the role of nuclear Cdk5 in the regulation of activity-dependent gene transcription and dendritic growth.
Keywords: dendrite, nuclear translocation, transcription, BDNF, neurotrophin, MeCP2
Introduction
Neuronal information is received, integrated, and processed at the dendrites, the functioning of which depends on their proper growth and patterning. Although the early establishment of the specific patterns of dendritic trees in different types of neurons is determined intrinsically by genetic programs, the growth and refinement of dendritic branches are modulated by neuronal activity (Katz and Shatz, 1996; Konur and Ghosh, 2005). Blocking neuronal activity by tetrodotoxin in cultured neurons or intact animals leads to dendritic growth deficits. In contrast, membrane depolarization of cultured neurons by KCl or increasing synaptic activity by exposing experimental animals to an enriched environment promotes dendritic growth (McAllister, 2000). Activity-dependent dendritic growth requires gene transcription. Among activity-dependent genes, bdnf, which encodes brain-derived neurotrophic factor (BNDF), is one of the most extensively studied. BDNF plays important roles in dendrite development and synaptic plasticity (Huang and Reichardt, 2001) and its transcription is substantially upregulated by neuronal activity in cultured neurons in vitro and by sensory experience in vivo (Barth, 2007).
Although the importance of activity-dependent gene transcription in dendrite development is well documented, how signals generated by neuronal activity are transduced into the nucleus to regulate transcription has only recently begun to be elucidated. Several transcription factors are implicated in mediating activity-dependent gene transcription crucial for dendritic growth. The cAMP response element-binding protein (CREB) plays a central role in long-term synaptic plasticity and memory in different animals including Drosophila, Aplysia, and mice (Alberini, 2009). CREB is also implicated in mediating the activity-dependent expression of BDNF during cortical development (Hong et al., 2008). In addition, the calcium-dependent transcription factors NeuroD and CREST are crucial for membrane depolarization-induced gene expression and dendritic growth during early development (Aizawa et al., 2004; Gaudillière et al., 2004). Recent studies also provide important insights into how these transcription factors are activated by neuronal activity, which involves posttranslational modifications such as phosphorylation. For example, CREB is activated upon phosphorylation by the kinase CaMKIV after neuronal depolarization. CREB is also activated by a slower and more persistent mechanism that involves the nuclear translocation of MAPK and PKA (Deisseroth et al., 2003).
Cortical pyramidal neurons of mice lacking cyclin-dependent kinase 5 (Cdk5) exhibit abnormal dendritic morphology (Nikolic et al., 1996; Ohshima et al., 2007). We have reported previously that Cdk5 mediates BDNF-induced dendritic growth in hippocampal neurons through the phosphorylation of BDNF receptor (TrkB) and the regulation of actin dynamics (Cheung et al., 2007). Although Cdk5 plays an essential role in BDNF-induced dendrite development, whether and how Cdk5 participates in activity-dependent brain development remains unclear. In addition, given the nuclear localization of Cdk5 in neurons (Ino and Chiba, 1996) and the various transcriptional regulators (e.g., STAT3, MEF2, and mSds3) as Cdk5 substrates (Su and Tsai, 2011), it is tempting to speculate that Cdk5 regulates dendrite development through transcriptional regulation in conjunction with cytoskeletal reorganization. Accordingly, the present study determines whether and how nuclear Cdk5 promotes neuronal activity-dependent dendrite development in neurons.
Here, we show that Cdk5 translocates into the nucleus after neuronal depolarization. Moreover, loss of Cdk5 significantly reduces dendritic growth and activity-induced bdnf transcription and the dendritic defects can be rescued by wild-type Cdk5, but not the nuclear-localization-deficient mutant. Intriguingly, we found that Cdk5 function is mediated through the regulation of the phosphorylation—and thus the activity of—methyl-CpG-binding protein 2 (MeCP2), a key transcriptional repressor. Therefore, the present study reveals a novel mechanism underlying activity-dependent dendritic growth that involves the nuclear translocation of Cdk5 and its subsequent regulation of the transcriptional program.
Materials and Methods
Antibodies and constructs.
Antibodies against Cdk5 (C-8, DC-17) and Lamin B (C-7) were from Santa Cruz Biotechnology; antibodies against actin (A3853), FLAG (M2), and MeCP2 (M6818) were from Sigma-Aldrich; MeCP2 antibody (ab2828) was from Abcam; proline-directed phosphoserine antibody was purchased from Cell Signaling Technology; and phospho-S421-MeCP2 antibody was a kind gift from Prof. Michael Greenberg (Harvard Medical School). The target sequence of rat Cdk5 shRNA used in this study was 5′-CCGGGAGATCTGTCTACTC-3′. The GFP-Cdk5 and NES-GFP-Cdk5 constructs were kind gifts from Prof. Karl Herrup (The Hong Kong University of Science and Technology). The nuclear export signal (NES)-Cdk5 was generated by PCR using the MAPKK NES (ALQKKLEELELD).
Protein extraction and fractionation.
The visual cortices of mice of either sex were collected as described previously (Yoshii et al., 2013). Cytosolic and nuclear fractionation was performed using the Nuclear/Cytosol Extraction Kit (BioVision) and the Nuclear Complex Co-IP Kit (Active Motif).
RNA extraction, ChIP, and quantitative real-time PCR.
RNA was extracted using the QIAGEN RNA extraction kit according to the manufacturer's instructions. For ChIP, cells were lysed and fragmented with a Covaris S220 focused ultrasonicator, followed by immunoprecipitation with rabbit polyclonal MeCP2 antibody. Quantitative real-time PCR was performed with fast-standard SYBR green dye using an AB7500 real-time PCR machine as described previously (Ng et al., 2013). The following primers were used for real-time PCR: bdnf exon IV forward 5′-CTGCCTTGATGTTTACTTTGACAAG-3′, bdnf exon IV reverse 5′-ACCATAGTAAGGAAAAGGATGGTGAT-3′; bdnf forward 5′-TTGAGCACGTGATCGAAGAG-3′, bdnf reverse 5′-CCAGCAGAAGAGCAGAGGA-3′; bdnf exon IV (promotor) forward 5′-GCGCGGAATTCTGATTCTGGTAA T-3′; bdnf exon IV (promotor) reverse 5′-GAGAGGGCTCCACGCTGCCTTGAC G-3′; hprt1 mRNA (endogenous control) forward 5′-TGACACTGGTAAAACAATGCA-3′, reverse 5′-GGTCCTTTTCACCAGCAAGCT-3′.
Cell culture and transfection.
Primary hippocampal or cortical neurons were prepared from embryonic day 18 (E18) rats or transgenic mice and maintained in neurobasal medium with 2% B27 supplement. Primary neurons were transfected using the calcium phosphate method as described previously (Goff et al., 2007) or were transfected with the Lipofectamine 2000 transfection kit according to the manufacturer's instructions.
For KCl depolarization, cells were depolarized with 50 mm KCl by adding 31% depolarization buffer (210 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, pH 7.4) to the culture medium (Flavell et al., 2006). For the control groups, KCl was substituted with the same concentration of NaCl. For the gene expression experiments, cells were pretreated with 1 μm TTX and 100 μm APV for 2 h.
Microarray and data analysis.
Cortical neurons at 10 d in vitro (DIV) were cultured from E18 Cdk5-knock-out embryos and littermates. RNA was analyzed with the GeneChip WT PLUS Reagent Kit (WT PLUS Kit; Affymetrix) according to the manufacturer's instructions. The microarray data were analyzed using R language (http://www.r-project.org/). Genes with p values <0.05 and changes of expression exceeding 1.5-fold between the experimental and control conditions (i.e., NaCl vs KCl or cdk5+/+ vs cdk5−/−) were selected to generate the heat map. Gene list analysis was performed using the PANTHER classification system (http://www.pantherdb.org/).
Confocal imaging and quantification.
Images were captured with Nikon A1 confocal microscopes with 40× oil-immersion lenses. The number and length of dendrites were quantified using ImageJ with the NeuronJ plugin (Meijering et al., 2004). Sholl analysis, which measures the number of intersections of neuronal dendrites crossing a series of concentric circles from the cell body, was performed using ImageJ embedded with the Sholl analysis plugin (A. Ghosh, University of California San Diego). Approximately 20–40 neurons from three independent experiments were measured.
Results
Cdk5 nuclear translocation is induced by neuronal activity
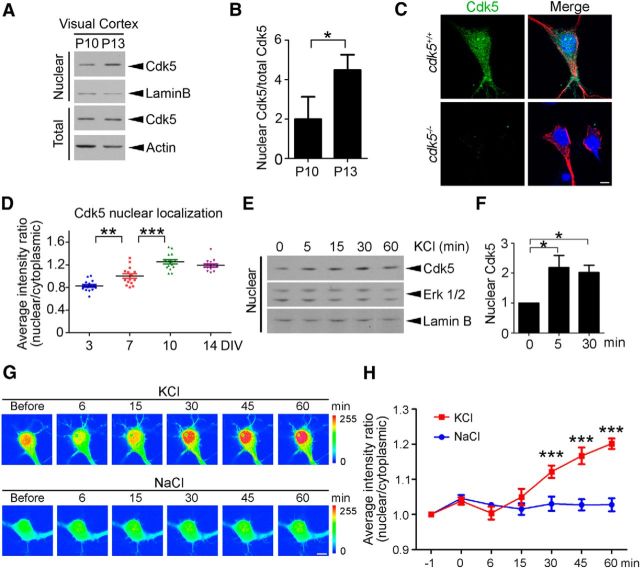
To determine whether Cdk5 is involved in activity-dependent dendritic development, we initially investigated whether increased neuronal activity regulates the subcellular localization of Cdk5. To this end, we determined the subcellular localization of Cdk5 in the mouse visual cortex before and after eye opening, when the animals receive visual stimulation. Interestingly, accumulation of Cdk5 in the nuclear fraction of the visual cortex was detected after eye opening (Fig. 1A,B), indicating that Cdk5 translocates into the nucleus after increased neuronal activity. To verify the nuclear localization of endogenous Cdk5 in neurons, hippocampal neurons derived from cdk5−/− mice and their wild-type littermates were stained with antibody against Cdk5. The results showed that Cdk5 immunoreactivity was colocalized with the dendritic marker MAP2 and nuclear marker DAPI in cdk5+/+ neurons, whereas the signal was absent in the cdk5−/− neurons (Fig. 1C). These results indicate that Cdk5 is localized in both the cytoplasm and nucleus of neurons. Cdk5 increasingly accumulated in the nuclei of cultured hippocampal neurons from 3 to 10 DIV, which corresponds to the period critical for dendritic outgrowth and maturation (Fig. 1D). Furthermore, Cdk5 was enriched in the nuclear fraction of the cultured neurons after KCl-induced neuronal depolarization (Fig. 1E,F), whereas the total level of Cdk5 did not change during treatment (data not shown). Notably, time-lapse confocal imaging demonstrated that the nuclear translocation of GFP-Cdk5 in hippocampal neurons was specifically induced by KCl-induced neuronal depolarization but not the control (NaCl) (Fig. 1G,H). These findings collectively indicate that neuronal activity promotes the nuclear translocation of Cdk5.
Figure 1.
Cdk5 translocates into the nucleus as a result of visual stimulation during neural development. A, B, Nuclear levels of Cdk5 were increased in the mouse visual cortex after eye opening. A, Western blot analysis. P, Postnatal day. B, Quantification: nuclear and total Cdk5 levels were normalized to those of Lamin B and actin, respectively (mean ± SEM; n = 4 from 3 experiments; Student's t test). C, Cdk5 is localized in both the nucleus and cytoplasm. Cultured hippocampal neurons derived from cdk5+/+ and cdk5−/− mice were stained with the nuclear marker DAPI (blue), as well as antibodies against Cdk5 (green) and the dendritic marker MAP2 (red). Scale bar, 10 μm. D, Cdk5 nuclear accumulation increased during neuronal maturation (mean ± SEM; n = 15 from two experiments; one-way ANOVA with Tukey's post hoc test, **p < 0.01; ***p < 0.001). E, F, Cdk5 translocated into the nucleus by neuronal depolarization. Cortical neurons were treated with 50 mm KCl for different times and the nuclear fractions were collected. E, Western blot analysis; Lamin B was used as a loading control. F, Quantification (mean ± SEM; n = 3 from 3 experiments; one-way ANOVA with Tukey's post hoc test, *p < 0.05). G, H, Confocal time-lapse imaging analysis of hippocampal neurons expressing GFP-Cdk5 after KCl treatment. Hippocampal neurons (2 DIV) were transfected with GFP-Cdk5. Two days after transfection, neurons were treated with 50 mm KCl or NaCl. G, Representative images showing Cdk5 translocated into the nucleus after 50 mm KCl treatment. H, Quantification of the average intensity signal (mean ± SEM; n = 4; two-way ANOVA with Bonferroni post hoc test, treatment × time F(6,36) = 24.52, p < 0.001, treatment F(1,6) = 11.69, p < 0.05, time F(6,36) = 29.95, p < 0.001, ***p < 0.001). Scale bar, 10 μm.
Inhibition of Cdk5 attenuates activity-dependent dendrite development
Neuronal activity induces dendritic growth (Wong and Ghosh, 2002). Therefore, we investigated whether Cdk5 activity is required for neuronal depolarization-induced dendritic growth. Although depolarization by KCl increased the number of dendrites in cultured hippocampal neurons, treatment with the Cdk5 inhibitor roscovitine (Ros) significantly attenuated the induced number of dendrites (Fig. 2A,B). To confirm the essential role of Cdk5 in activity-dependent dendrite development, hippocampal neurons derived from cdk5+/+ and cdk5−/− mice were treated with KCl and activity-induced dendritic growth was examined. Neuronal depolarization significantly increased dendritic number, whereas the induction was abolished in cdk5−/− neurons (Fig. 2C,D). Therefore, Cdk5 is critical for the growth of dendritic arbors induced by neuronal depolarization.
Figure 2.
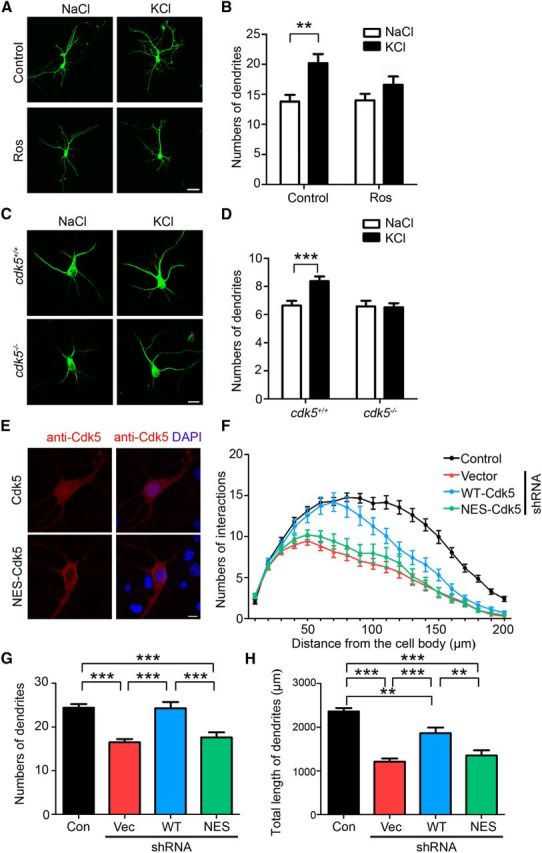
Cdk5 inhibition attenuates activity-dependent dendrite development. A, B, The increase in dendrite arborization after 24 h of KCl depolarization was attenuated by Ros. Cultured hippocampal neurons expressing GFP at 3 DIV were cotreated with 25 μm Ros. A, Representative images showing neuron morphology. Scale bar, 20 μm. B, Quantification of dendrite number (mean ± SEM; n = 10; two-way ANOVA with Bonferroni post hoc test, treatment × drug F(1,18) = 2.41, p = 0.1382, treatment F(1,18) = 1.93, p = 0.1821, drug F(1,18) = 11.48, p < 0.01, **p < 0.01). C, D, The KCl-induced dendritic growth was abolished in cdk5−/− neurons. C, Cultured hippocampal neurons from cdk5−/− mice and their littermates were treated with KCl and stained with dendritic marker MAP2 (green). Scale bar, 10 μm. D, Quantification of the dendrite (mean ± SEM; n = 6; two-way ANOVA with Bonferroni post hoc test, treatment × genotype F(1,10) = 14.09, p < 0.001, treatment F(1,10) = 12.02, p < 0.01, genotype F(1,10) = 5.25, p < 0.05, ***p < 0.001). E–H, Cdk5 nuclear expression is important for dendrite development. E, Subcellular expressions of WT-Cdk5 and NES-Cdk5 in cultured hippocampal neurons. Cultured hippocampal neurons transfected with Cdk5 or its nuclear localization-deficient mutant (NES-Cdk5) were stained with Cdk5 antibody. Scale bar, 10 μm. F–H, Hippocampal neurons (7 DIV) were transfected with Cdk5 shRNA together with RNAi-resistant Cdk5 constructs. F, Sholl analysis of transfected neurons with Cdk5 shRNA and Cdk5 constructs. G, H, Knock-down of Cdk5 expression significantly reduced dendrite number and length. Coexpression of wild-type Cdk5 but not NES-Cdk5 rescued the defective dendritic arborization (mean ± SEM; n = 30 from three experiments; one-way ANOVA with Tukey's post hoc test, **p < 0.01, ***p < 0.001).
Nuclear localization of Cdk5 is required for dendrite development in hippocampal neurons
Given that Cdk5 is required for activity-induced dendritic growth and accumulates in the nucleus as a result of increased neuronal activity, we hypothesized that the nuclear localization of Cdk5 plays an important regulatory role during dendrite development. To test this hypothesis, a nuclear-localization-deficient Cdk5 mutant was constructed by adding an NES (Zhang et al., 2010), which reduced the nuclear expression of Cdk5 (Fig. 2E). Cdk5 knock-down in hippocampal neurons by RNAi significantly reduced dendrite number and length. Importantly, coexpression of wild-type Cdk5 partially rescued the defective dendritic arborization caused by the Cdk5 RNAi, but the nuclear localization-deficient mutant (NES-Cdk5) failed to rescue the dendritic defects (Fig. 2F–H). These results suggest that the nuclear localization of Cdk5 is required for its function in dendrite development.
Genome-wide analysis of Cdk5-regulated activity-dependent genes
To investigate how Cdk5 promotes activity-dependent dendrite development, an unbiased microarray analysis was performed to identify activity-dependent genes differentially regulated in cortical neurons derived from wild-type versus Cdk5-knock-out embryos. First, we identified that, upon KCl treatment, 2973 genes (including c-Fos, Arc, bdnf, and Npas4) were upregulated and 3230 genes were downregulated (KCl vs NaCl, data not shown); many of these genes have been identified in previous studies, suggesting robust methodology (Lin et al., 2008). Among the activity-regulated genes, 332 were significantly altered in cdk5−/− neurons compared with cdk5+/+ neurons after KCl treatment (cdk5−/− vs cdk5+/+ under KCl condition; Fig. 3A,B; Gene Expression Omnibus accession number GSE68320). Among these, 135 and 139 genes were downregulated and upregulated in cdk5−/− neurons, respectively (Fig. 3A,B). These results indicate that Cdk5 can regulate transcription during membrane depolarization in a bidirectional manner, as a transcriptional activator, or as a transcriptional repressor.
Figure 3.
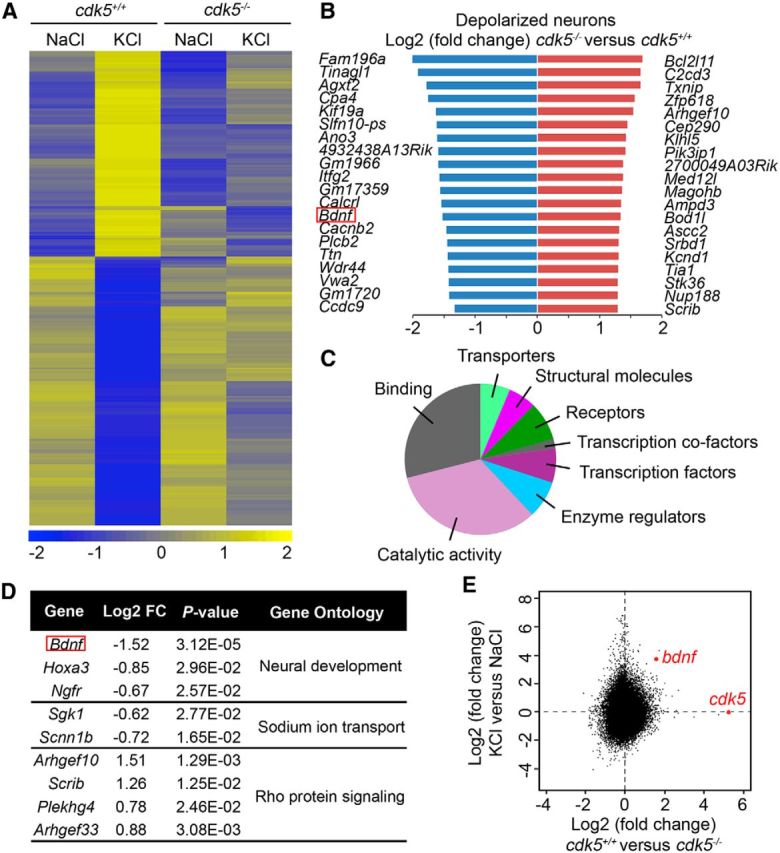
Cdk5 is essential for activity-dependent transcriptional regulation. A, Heat map showing the clustering of 388 activity-dependent probe sets (representing 332 unique genes) that are differentially expressed between cdk5+/+ and cdk5−/− cortical neurons upon KCl treatment (n = 3 mice per group). Cortical neurons (10 DIV) derived from cdk5+/+ and cdk5−/− mice were treated with 50 mm KCl or NaCl for 6 h to examine the mRNA expression. The expression level of each probe set is normalized to a mean of 0 and SD of 1 (log2). Expression values are displayed within the range [−2, 2], with levels above and below the mean displayed in yellow and blue, respectively. B, List of the top 20 genes showing the differentially expressed genes (upregulated and downregulated genes are shown in red and blue, respectively) in cdk5−/− neurons relative to cdk5+/+ neurons after KCl depolarization. C, Biological functions of putative Cdk5-regulated genes on the basis of gene ontology information provided by the PANTHER classification system. D, Downregulated and upregulated genes that may function in dendrite development on the basis of the DAVID database (https://david.ncifcrf.gov/). E, Comparison of the expression levels of each probe set between cdk5+/+ and cdk5−/− neurons (x-axis) after KCl depolarization (y-axis). Two probe sets for cdk5 and bdnf are denoted in red, respectively.
Gene function analysis (Mi et al., 2013) demonstrated that Cdk5 regulates a wide variety of genes such as those encoding cell surface receptors and their ligands and transporters, enzymes such as kinases and phosphatases, transcriptional factors and cofactors, and genes involved in cytoskeleton-modulating pathways (Fig. 3C). Although some Cdk5-regulated genes (e.g., bdnf, Cdh1, and Cacnb2) are implicated in neuronal development (Van Aelst and Cline, 2004; Tan et al., 2010; Breitenkamp et al., 2014), the functions of most of the genes identified in this analysis in dendrite development are uncharacterized. Interestingly, among the activity-induced genes, bdnf was one of the most significantly downregulated in cdk5−/− neurons (Fig. 3D,E).
Cdk5 regulates activity-dependent gene expression through the modulation of MeCP2 transcriptional activity
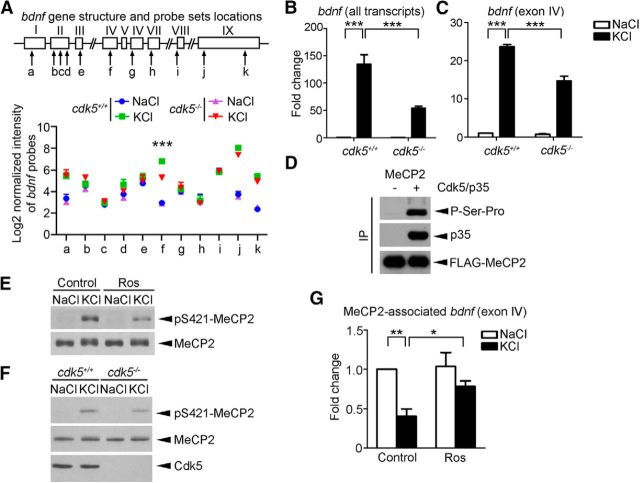
The activity-dependent gene bdnf promotes dendritic growth both in vitro and in vivo (Van Aelst and Cline, 2004). Notably, the bdnf transcript that contains exon IV promoter (induced by activity) was consistently decreased in the absence of Cdk5 in the KCl condition in microarray analysis (>2-fold; Fig. 4A). Therefore, we examined the activity-induced changes of bdnf transcripts in cdk5+/+ and cdk5−/− cortical neurons triggered by membrane depolarization using quantitative real-time PCR. Although treatment with KCl markedly upregulated the expressions of all bdnf transcripts and the exon IV transcript, the increase of their respective mRNA levels was attenuated in cdk5−/− neurons compared with cdk5+/+ neurons (Fig. 4B,C). These results suggest that Cdk5 specifically regulates activity-dependent bdnf expression.
Figure 4.
Cdk5 regulates MeCP2 phosphorylation and DNA-binding activity. A, Schematic diagram of mouse bdnf; exons and introns are represented by boxes and lines, respectively. The probes used are denoted as a–k. The log2 values of the normalized intensity of individual probe set are plotted (mean ± SEM from three independent experiments; ***p < 0.005, cdk5−/− vs cdk5+/+ in the KCl condition, one-way ANOVA with Tukey's post hoc test). B, C, Fold changes of all bdnf transcripts and activity-induced exon IV transcript in cdk5+/+ and cdk5−/− cortical neurons triggered by membrane depolarization determined by quantitative real-time PCR (mean ± SEM; n = 3; one-way ANOVA with Tukey's post hoc test, ***p < 0.001). Cortical neurons (10 DIV) derived from cdk5+/+ and cdk5−/− mice were treated with 50 mm KCl or NaCl for 6 h to examine bdnf mRNA expression. D, MeCP2 was phosphorylated by Cdk5/p35 at Ser-Pro site(s) in HEK293T cells. FLAG-tagged MeCP2 was immunoprecipitated by anti-FLAG antibody, followed by Western blotting with antibody against proline-directed phosphoserine antibody. E, F, Regulation of KCl-induced Ser421 MeCP2 phosphorylation by Cdk5. E, Cultured cortical neurons were pretreated with 25 μm DMSO control or Ros for 1 h, followed by 50 mm KCl or NaCl for 30 min. The experiment was repeated at least three times. F, KCl-induced Ser421 MeCP2 phosphorylation was attenuated in cdk5−/− neurons. G, ChIP assay of MeCP2 followed by real-time PCR using bdnf exon IV promoter-specific primers (data were normalized to those of the Control+NaCl group; mean ± SEM; n = 5; one-way ANOVA with Tukey's post hoc test, *p < 0.05, **p < 0.01). Cultured cortical neurons were pretreated with 25 μm DMSO control or Ros for 1 h, followed by 50 mm KCl or NaCl for 90 min.
Activity-dependent bdnf transcription was recently reported to be controlled by MeCP2, mutations of which cause the neurological disorder Rett syndrome (Chen et al., 2001; Guy et al., 2001; Shahbazian et al., 2002). Because our previous mass spectrometry screening identified MeCP2 as a potential substrate of Cdk5 and because the activity of nuclear Cdk5 was elevated by membrane depolarization (data not shown), we investigated whether Cdk5 can phosphorylate MeCP2 and regulate its transcriptional repressor activity. Using antibodies that specifically recognize proline-directed phosphoserine residues, we found that Cdk5/p35 coexpression specifically induced MeCP2 serine phosphorylation (Fig. 4D), but not threonine phosphorylation (data not shown). MeCP2 is reported to be phosphorylated at Ser421 by CaMKII in response to neuronal activity and its specific phosphorylation is critical for activity-dependent bdnf transcription and dendrite development (Zhou et al., 2006). Intriguingly, the KCl-induced Ser421 phosphorylation of MeCP2 was attenuated by Ros pretreatment and reduced in cdk5−/− neurons (Fig. 4E,F). Therefore, the results indicate that Cdk5 is required for the activity-dependent phosphorylation of MeCP2 at Ser421. We subsequently investigated whether Cdk5 modulates the transcriptional repressor activity of MeCP2. ChIP assay revealed that membrane depolarization reduced the association of MeCP2 with bdnf exon IV by ∼50% in control neurons upon KCl stimulation (Fig. 4G). In contrast, this release of MeCP2 from bdnf exon IV was significantly attenuated after Ros treatment. These results suggest that Cdk5 activity is required for the effective release of MeCP2 from the bdnf promoter and the subsequent transcription of bdnf upon neuronal activity.
Discussion
Proper dendrite growth and branching are crucial for neural circuit formation and nervous system function. Although defects in dendrite development are associated with various human mental disorders such as autism and schizophrenia (Kaufmann and Moser, 2000), relatively little is known about the molecular mechanisms that control dendrite development. The present study identified Cdk5-MeCP2-dependent transcriptional regulation as an important signaling axis in activity-dependent dendrite development. Although Cdk5 regulates transcription and dendritic growth through the phosphorylation, thus modulating the activity of MeCP2, a key transcriptional repressor for activity-regulated genes, this regulatory mechanism is precisely controlled by the activity-dependent nuclear translocation of Cdk5. The present findings advance the current understanding of how neuronal activity shapes dendritic arborization patterns and connectivity during nervous system development.
The formation of dendritic arbors is tightly regulated by neuronal activity during neural development and one of the major underlying mechanisms involves the expressions of activity-dependent genes such as that of the neurotrophin BDNF (Katz and Shatz, 1996; Konur and Ghosh, 2005). Although gene transcription can be specifically regulated by the phosphorylation of transcriptional regulators, how protein kinases propagate signals generated by neuronal activity to the transcriptional machinery remains poorly understood. Despite being studied extensively as a key kinase that regulates cytoskeleton in brain development and synaptic plasticity (Su and Tsai, 2011), the roles of Cdk5 in regulating transcriptional machinery have only begun to be elucidated. The present findings show that Cdk5 transduces the extracellular signal to the nucleus to regulate activity-induced gene transcription for the growth of dendritic arbors, which may serve as an important mechanism that molds the neural circuit in response to sensory experience during brain development.
Various transcriptional regulators, including MEF2 and mSds3, are substrates of Cdk5. Cdk5 phosphorylates and consequently inhibits MEF2 transcriptional activity, suggesting that Cdk5 might regulate the development of dendritic spines and excitatory synapses via the regulation of MEF2-mediated gene expression (Flavell et al., 2006; Pulipparacharuvil et al., 2008). Moreover, we reported previously that Cdk5 phosphorylates mSds3, a corepressor of mSin3-histone deacetylase (HDAC), so this phosphorylation is crucial for its repressive activity (Li et al., 2004). Notably, HDAC is implicated in the activity-regulated expression of bdnf (Zhang et al., 2007). Furthermore, other transcription factors that mediate activity-dependent gene transcription, such as NeuroD, SRF, ELK, NFAT, CREST, and NF-κB, also contain putative Cdk5 phosphorylation sites (Z. Liang and N. Y. Ip, unpublished observations); however, whether they are regulated by Cdk5 has not been investigated. In addition to the phosphorylation of transcription factors, Cdk5 may regulate the transcriptional machinery through the modulation of different signaling pathways such as the cAMP/CREB pathway (Guan et al., 2011). Our finding of the nuclear translocation of Cdk5 upon activity stimulation provides a molecular basis for how this kinase modulates transcription.
The results of our microarray analysis are the first to demonstrate that Cdk5 is required for the activation and repression of distinct subsets of activity-regulated genes during membrane depolarization (Fig. 3A). Among the genes upregulated by neuronal activity, the expression of bdnf exon IV promoter-specific transcript was markedly attenuated in Cdk5-deficient neurons (Fig. 4A–C). Indeed, knock-in mice with a mutation in bdnf exon IV exhibit defects in dendrite morphology (Hong et al., 2008). These lines of evidence collectively highlight the significance of Cdk5 in activity-induced bdnf transcription, which is critical for dendrite development.
How does Cdk5 regulate activity-dependent bdnf gene transcription? BDNF is encoded by a complex gene with multiple distinct promoters that give rise to at least nine transcript variants (Aid et al., 2007). In the absence of neuronal activity, MeCP2, a key contributor to Rett syndrome, inhibits bdnf gene transcription by selectively binding to its exon IV promoter. Membrane depolarization triggers the calcium-dependent Ser421 phosphorylation of MeCP2, thereby releasing MeCP2 from bdnf exon IV and activating bdnf gene transcription (Chen et al., 2003; Zhou et al., 2006). In the present study, the results of a phosphorylation assay identified MeCP2 as a novel substrate of Cdk5. Inhibition of Cdk5 attenuated activity-induced Ser421 MeCP2 phosphorylation in cortical neurons and impaired its effective release from bdnf. Notably, MeCP2 Ser421 phosphorylation may facilitate a genome-wide response of chromatin to neuronal activity (Cohen et al., 2011). Given that the structure of chromatin modulates the access of condensed genomic DNA to the regulatory transcription machinery proteins, thereby controlling gene expression, it would be of interest to further characterize the physiological functions of the Cdk5-dependent regulation of MeCP2 in experience-dependent dendrite development and remodeling; the results of such a study may shed light on the molecular basis of normal brain development and the etiology of neurological disorders associated with dendrite abnormalities, such as autism and schizophrenia.
Footnotes
We are grateful to Prof. Michael Greenberg (Harvard Medical School) for sharing MeCP2 antibodies and Prof. Karl Herrup (The Hong Kong University of Science and Technology) for the GFP-Cdk5 and NES-GFP-Cdk5 constructs. We also thank Cara Kwong, Busma Butt, William Chau, Renfei Wu, Yilei Cai, and Dr. Maggie Chu for their excellent technical assistance as well as Prof. Tom Cheung and other members of the Ip laboratory for helpful discussions. This study was supported in part by the Research Grants Council of Hong Kong SAR (HKUST660810, HKUST661111, HKUST661212, and HKUST660213), the National Key Basic Research Program of China (2013CB530900), Hong Kong Research Grants Council Theme-based Research Scheme (T13-607/12R), and the SH Ho Foundation.
The authors declare no competing financial interests.
References
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, Gurevich I, Cowan M, Ghosh A. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. doi: 10.1126/science.1089845. [DOI] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL. Visualizing circuits and systems using transgenic reporters of neural activity. Curr Opin Neurobiol. 2007;17:567–571. doi: 10.1016/j.conb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitenkamp AF, Matthes J, Nass RD, Sinzig J, Lehmkuhl G, Nürnberg P, Herzig S. Rare mutations of CACNB2 found in autism spectrum disease-affected families alter calcium channel function. PLoS One. 2014;9:e95579. doi: 10.1371/journal.pone.0095579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cheung ZH, Chin WH, Chen Y, Ng YP, Ip NY. Cdk5 is involved in BDNF-stimulated dendritic growth in hippocampal neurons. PLoS Biol. 2007;5:e63. doi: 10.1371/journal.pbio.0050063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE, Greenberg ME. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Mermelstein PG, Xia H, Tsien RW. Signaling from synapse to nucleus: the logic behind the mechanisms. Curr Opin Neurobiol. 2003;13:354–365. doi: 10.1016/S0959-4388(03)00076-X. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Gaudillière B, Konishi Y, de la Iglesia N, Yao GI, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/S0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Goff MJ, Johnson RN, McDonald HR, Ai E, Jumper JM, Fu A. Intravitreal bevacizumab for previously treated choroidal neovascularization from age-related macular degeneration. Retina. 2007;27:432–438. doi: 10.1097/IAE.0b013e318042b53f. [DOI] [PubMed] [Google Scholar]
- Guan JS, Su SC, Gao J, Joseph N, Xie Z, Zhou Y, Durak O, Zhang L, Zhu JJ, Clauser KR, Carr SA, Tsai LH. Cdk5 is required for memory function and hippocampal plasticity via the cAMP signaling pathway. PLoS One. 2011;6:e25735. doi: 10.1371/journal.pone.0025735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ino H, Chiba T. Intracellular localization of cyclin-dependent kinase 5 (CDK5) in mouse neuron: CDK5 is located in both nucleus and cytoplasm. Brain Res. 1996;732:179–185. doi: 10.1016/0006-8993(96)00523-9. [DOI] [PubMed] [Google Scholar]
- Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Konur S, Ghosh A. Calcium signaling and the control of dendritic development. Neuron. 2005;46:401–405. doi: 10.1016/j.neuron.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Li Z, David G, Hung KW, DePinho RA, Fu AK, Ip NY. Cdk5/p35 phosphorylates mSds3 and regulates mSds3-mediated repression of transcription. J Biol Chem. 2004;279:54438–54444. doi: 10.1074/jbc.M411002200. [DOI] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK. Cellular and molecular mechanisms of dendrite growth. Cereb Cortex. 2000;10:963–973. doi: 10.1093/cercor/10.10.963. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng YP, Chen Y, Hu Y, Ip FC, Ip NY. Olean-12-eno[2,3-c] [1,2,5]oxadiazol-28-oic acid (OEOA) induces G1 cell cycle arrest and differentiation in human leukemia cell lines. PLoS One. 2013;8:e63580. doi: 10.1371/journal.pone.0063580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M, Dudek H, Kwon YT, Ramos YF, Tsai LH. The cdk5/p35 kinase is essential for neurite outgrowth during neuronal differentiation. Genes Dev. 1996;10:816–825. doi: 10.1101/gad.10.7.816. [DOI] [PubMed] [Google Scholar]
- Ohshima T, Hirasawa M, Tabata H, Mutoh T, Adachi T, Suzuki H, Saruta K, Iwasato T, Itohara S, Hashimoto M, Nakajima K, Ogawa M, Kulkarni AB, Mikoshiba K. Cdk5 is required for multipolar-to-bipolar transition during radial neuronal migration and proper dendrite development of pyramidal neurons in the cerebral cortex. Development. 2007;134:2273–2282. doi: 10.1242/dev.02854. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, Greengard P, Nairn AC, Nestler EJ, Cowan CW. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong D, Paylor R, Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/S0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Su SC, Tsai LH. Cyclin-dependent kinases in brain development and disease. Annu Rev Cell Dev Biol. 2011;27:465–491. doi: 10.1146/annurev-cellbio-092910-154023. [DOI] [PubMed] [Google Scholar]
- Tan ZJ, Peng Y, Song HL, Zheng JJ, Yu X. N-cadherin-dependent neuron-neuron interaction is required for the maintenance of activity-induced dendrite growth. Proc Natl Acad Sci U S A. 2010;107:9873–9878. doi: 10.1073/pnas.1003480107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aelst L, Cline HT. Rho GTPases and activity-dependent dendrite development. Curr Opin Neurobiol. 2004;14:297–304. doi: 10.1016/j.conb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Yoshii A, Zhao JP, Pandian S, van Zundert B, Constantine-Paton M. A Myosin Va mutant mouse with disruptions in glutamate synaptic development and mature plasticity in visual cortex. J Neurosci. 2013;33:8472–8482. doi: 10.1523/JNEUROSCI.4585-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li H, Herrup K. Cdk5 nuclear localization is p27-dependent in nerve cells: implications for cell cycle suppression and caspase-3 activation. J Biol Chem. 2010;285:14052–14061. doi: 10.1074/jbc.M109.068262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Kohlhaas M, Backs J, Mishra S, Phillips W, Dybkova N, Chang S, Ling H, Bers DM, Maier LS, Olson EN, Brown JH. CaMKIIdelta isoforms differentially affect calcium handling but similarly regulate HDAC/MEF2 transcriptional responses. J Biol Chem. 2007;282:35078–35087. doi: 10.1074/jbc.M707083200. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]