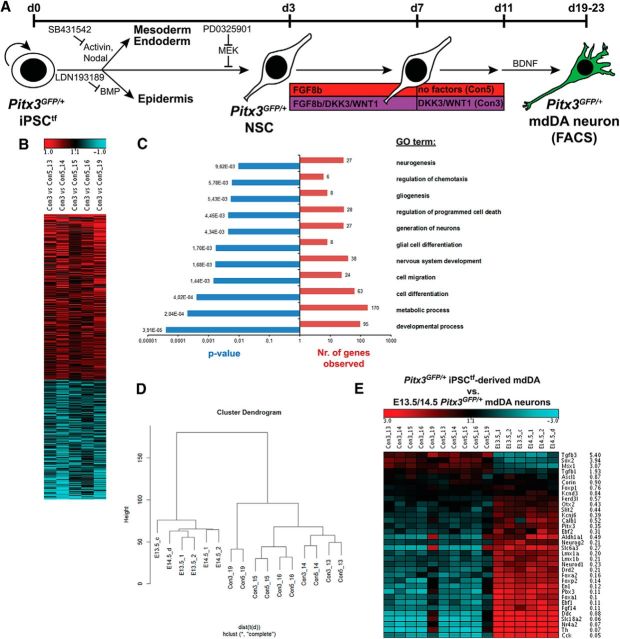
Figure 7.
Global gene expression changes in DKK3/WNT1-treated versus untreated iPSC-derived Pitx3GFP/+ mdDA neurons. A, Scheme of the monolayer protocol for the differentiation of Pitx3GFP/+ iPSCtfs with the conditions analyzed by microarray of FACS-sorted eGFP-expressing Pitx3GFP/+ mdDA neurons (5 independent differentiation experiments: 13, 14, 15, 16, and 19). B, C, Heat map (B) and GO term analysis (C) of differentially regulated genes in the DKK3 + WNT1-treated iPSC-derived Pitx3GFP/+ mdDA neurons (Con3) relative to the untreated cells (Con5) (red, upregulated; blue, downregulated genes). D, Hierarchical clustering analysis of the global gene expression profiles of E13.5/E14.5 primary and DKK3 + WNT1-treated (Con3) or untreated (Con5) iPSC-derived Pitx3GFP/+ mdDA neurons. Clustering was done with the R script hclust on probe sets with an average expression value >50 in at least one of the four groups. The ordinate in the hierarchical cluster dendrogram indicates the distance of samples. E, Expression levels of selected mdDA progenitor- or mdDA neuron-specific genes in DKK3 + WNT1-treated (Con3) and untreated (Con5) iPSC-derived Pitx3GFP/+ mdDA neurons, as well as their E13.5/E14.5 in vivo counterparts. Average ratios shown are Con3 (DKK3 + WNT1-treated) versus E13.5 Pitx3GFP/+ mdDA neurons.