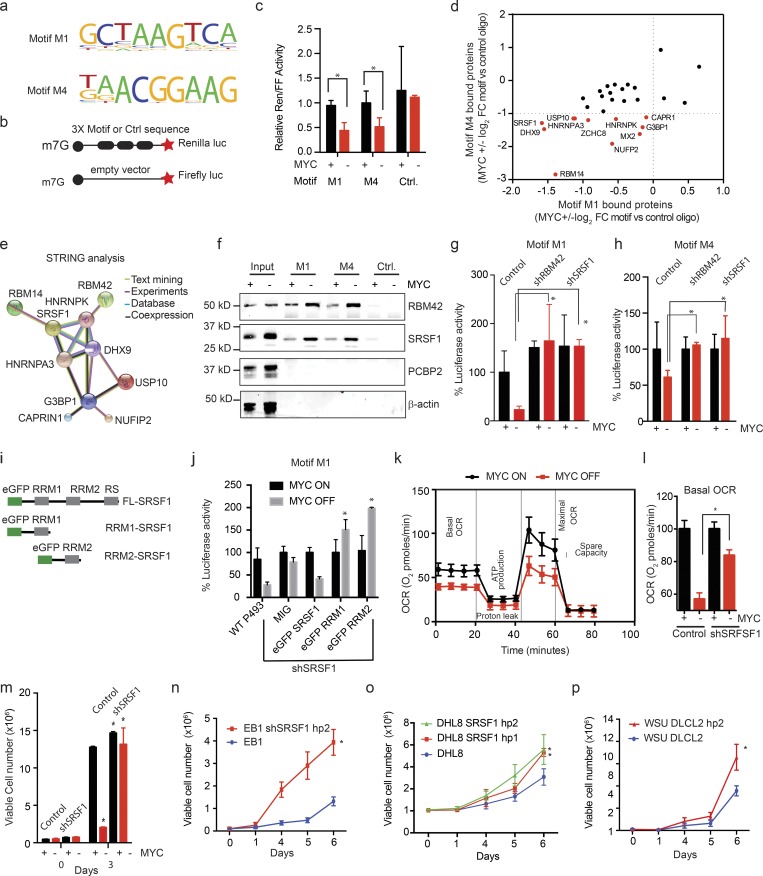
Figure 3.
MYC-dependent mRNAs have 5′UTR SRF1/RBM42 binding elements. (a) Motifs M1 and M4 are significantly enriched in MYC-dependent mRNAs and confer MYC sensitivity in reporter assays; motifs M2 and M3 are enriched and did not perform in the reporter assay (sequence in Table S2); n > 3 replicates. (b) Schematic of the translation reporter assay to test MYC responsiveness of 5′UTR motifs relative to a capped firefly luciferase reporter construct of equal length and GC content. (c) Translation reporter comparing the ratio of Renilla luciferase expressed under control of a 5′UTR with three repeats of the indicated motif and a capped firefly luciferase reporter construct of equal length and GC content in P493-6 cells under MYC ON and OFF (mean ± SD; n = 3 biological replicates, performed as five independent experiments). P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05. (d) Mass-spectrometric identification of proteins with MYC-dependent binding to motifs M1 and M4 compared with random RNA sequence (fold change [FC] of proteins showing >5 peptides detected at an FDR <0.1% are included); n = 1 biological sample in each group. FDR values were calculated using the original Benjamini and Hochberg method. (e) STRING analysis of interactions between proteins identified in mass spectrometry analysis shown in panel d. (f) Biotin pull down and immunoblot show MYC-dependent binding of RBM42 and SRSF1 to RNA oligomers encoding motif M1/M4 versus the control sequence in P493-6 cells; PCBP2 and β-actin are controls. n = 3 biological replicates. (g and h) Effect of knockdown of SRSF1 or RBM42 versus control shRNA on MYC-dependent luciferase translation controlled by motif M1 (g) or motif M4 (h); assay as shown in panel b. Values are mean ± SD. n > 3 replicates, representative data are shown from three independent experiments. (i) Schematic of wild-type and RRM-deleted versions of SRSF1 expression constructs fused with enhanced GFP (eGFP). (j) Ectopic expression of wild-type SRSF1 could rescue the luciferase activity mediated by motif M1 in SRSF1 knockdown P493 cells while the RRM1- and RRM2-deleted SRSF1 protein could not mediate the translation repression. n = 3 replicates, mean ± SD from three replicates, representative data are shown from five independent experiments. P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05. (k) Mitochondrial stress assay to measure oxygen consumption rate (OCR) in MYC ON and OFF P493 cells. n = 4 replicates, mean ± SD from four replicates. Representative data are shown from three independent experiments. (l) Basal OCR in P493 control and shSRSF1-stable cells treated with tetracycline (0.1 μg/ml; 24 h). n = 4 replicates, mean ± SD from four replicates. Representative data are shown from three independent experiments. P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05. (m–p) Cell proliferation assays in indicated lymphoma cell lines, MYC ON and OFF in control and shSRSF1 P493-6 cells (m), control and shSRSF1 EB1 (n), DHL8 (o), and WSU-DLCL2 cells (p). n = 3 replicates in each group, mean ± SD from three replicates. Representative data are shown from three independent experiments. P values were calculated using an unpaired Student’s t test: *, P ≤ 0.05. Ctrl., control.