Figure 4. A conserved fusion-specific requirement for NTN1 in OFC and palate development.
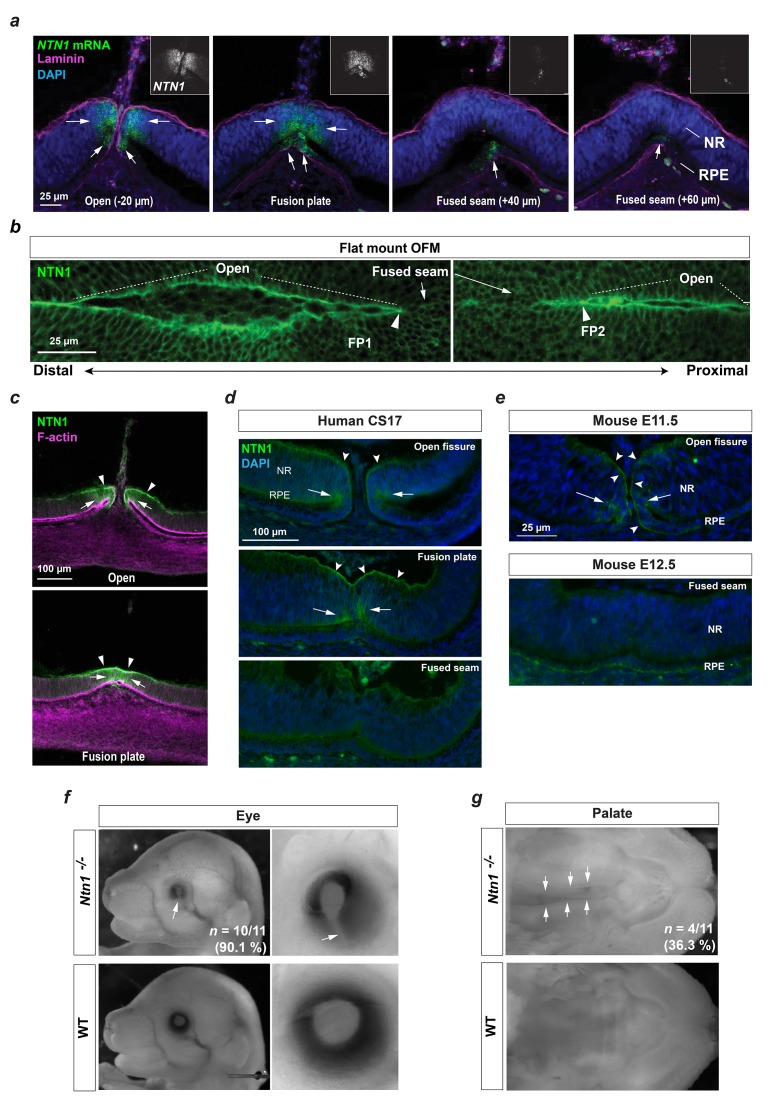
(a) RNAscope analysis of NTN1 mRNA (green, and grey in insets) in HH.St29 OFMs revealed fissure-specific NTN1 expression (arrows) with strongest signal observed at open regions and in the fusion plate, and reduced expression in the adjacently fused seam. NTN1 expression was localised to cells of both the NR and RPE. Fusion progression was indicated using anti-laminin co-immunofluorescence (magenta). Images shown are maximum intensity projections of confocal Z-stacks. (b) Single-plane confocal images of immunofluorescence analysis for NTN1 on flat-mounted distal (FP1) and proximal (FP2) OFM revealed enriched protein localisation at the edges of the open fissure margins and reduced in the fused seam. (c) Immunostaining on cryosectioned OFM at the open and fusion plate at HH.St29 revealed NTN1 was specifically localised to the basal lamina (arrowheads) and to the epithelia of the neural retina and RPE (arrows) at the OFM. (d) Immunostaining on CS17 human foetal eye sections revealed human Netrin-1 (hNTN1) was localised to NR epithelia (arrows) and at the overlying basal lamina (dented arrowheads) at the fissure margins. hNTN1 was absent from the fused seam epithelia. (e) Immunostaining for mouse Netrin-1 (mNtn1) in during active fusion stages (E11.5) showed mNtn1 was localised at the open fissure margins (arrow) in the basal lamina and to cells at the NR-RPE junction. mNtn1 was absent from this region in fused OFM seam at E12.5. (f) Ntn1-/- mice exhibited highly penetrant (~90%) bilateral coloboma (arrows; n = 10/11 homozygous E15.5-E16.5 animals analysed). (g) Cleft secondary palate (arrows) was observed in ~36% of Ntn1-/- embryos at E15.5-E16.5 (4/11 homozygous animals).
Figure 4—figure supplement 1. Developmental NTN1 expression profiling in chick eye and OF.
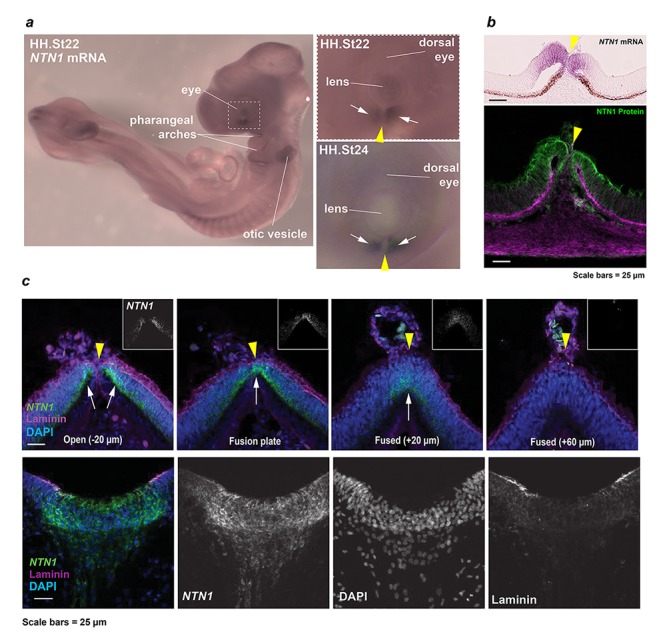
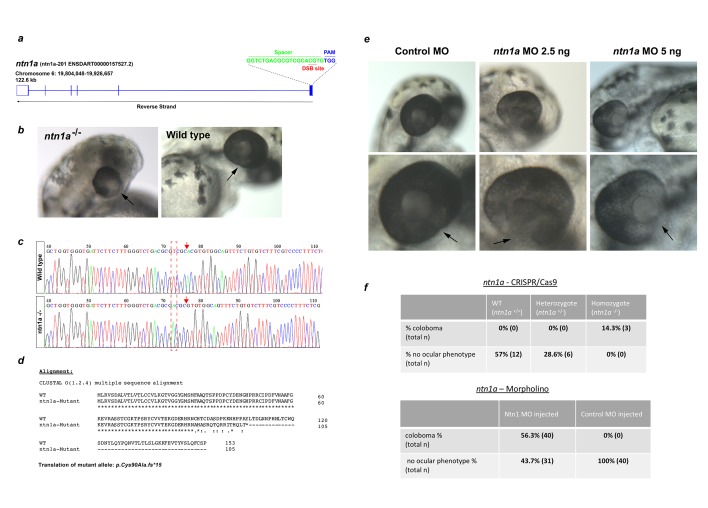
Figure 4—figure supplement 2. Analyses of mouse Ntn1 knockout fissures during fusion.
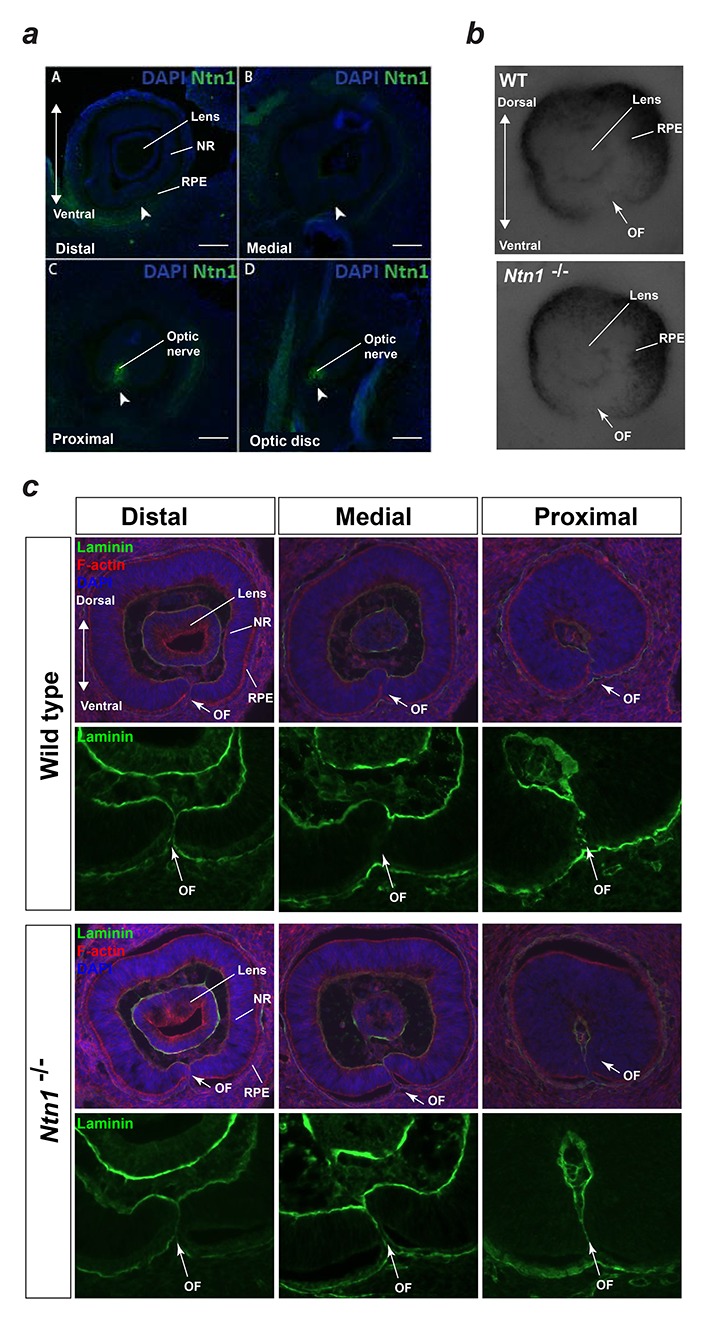
Figure 4—figure supplement 3. Gross ocular phenotype analyses of ntn1-deficient zebrafish.
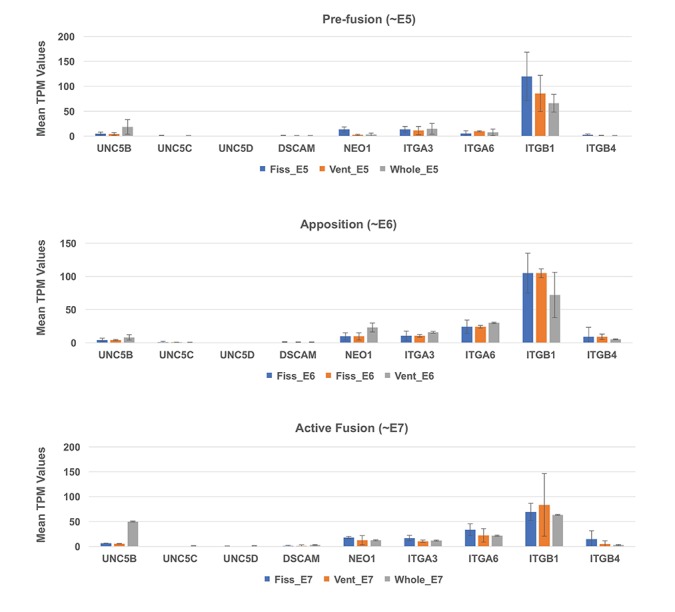
Figure 4—figure supplement 4. Expression profiling for known interactors of NTN1.