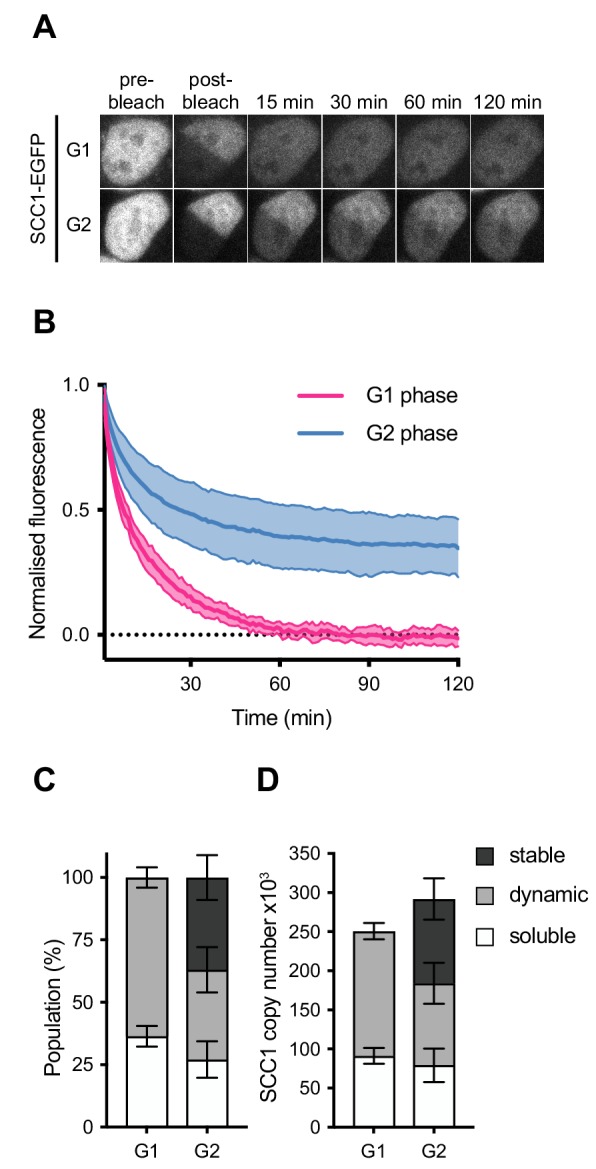
Figure 3. Dynamics and distribution of nuclear SCC1-mEGFP in G1 and G2 phase.
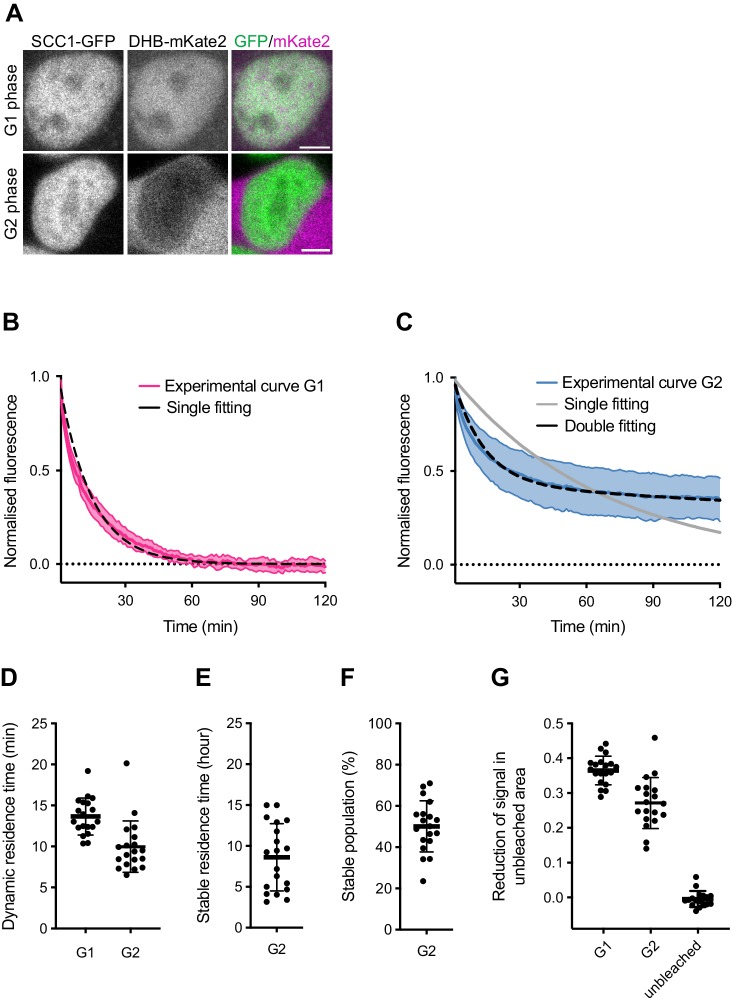
(A) Images of inverse fluorescence recovery after photobleaching (iFRAP) in SCC1-mEGFP cells in G1 and G2 phase (Figure 3—figure supplement 1A). Half of the nuclear SCC1-mEGFP fluorescent signal was photobleached and the mean fluorescence in the bleached and unbleached regions was monitored by time-lapse microscopy. (B) The difference in fluorescence signals between the bleached and unbleached regions was normalised to the first post bleach image and plotted (mean ± S.D., n = 19 per condition). (C) SCC1-mEGFP distribution in the nucleus in G1 and G2 phase. Dynamic and stable populations were estimated using curve fittings from Figure 3—figure supplement 1B,C. Soluble populations were estimated by measuring the reduction in signal intensity in the unbleached area after bleaching (Figure 3—figure supplement 1G). (D) FCS/FRAP estimates of soluble, dynamic and stable nuclear SCC1-mEGFP copy number (see Table 3 for exact values).