Figure 3.
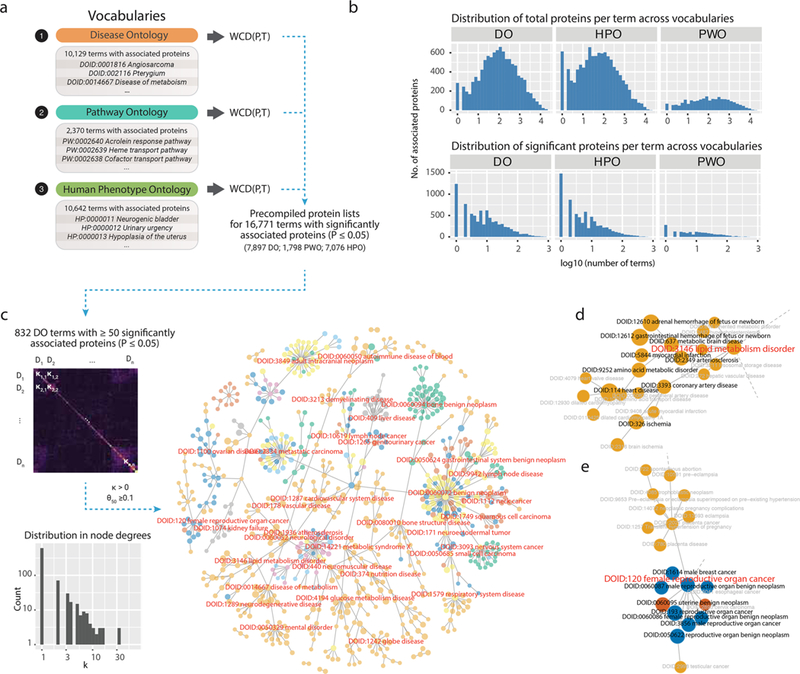
Prioritized proteins across the human diseasome. (a) Schematics for precompiling popular protein terms from three standard vocabularies related to human diseases and disease processes. (b) Distribution of total proteins per term in three vocabularies. The distribution of number of total (left) and significantly associated (right) proteins per term in each vocabulary (P ≤ 0.05). (c) Correlation matrix of protein associations for 832 Disease Ontology (DO) terms with 50 or more proteins associated at P ≤ 0.05. A minimal spanning tree of DO terms based on similarity of associated proteins. A protein network is constructed using all DO terms associated with any proteins as nodes. Edges connect pairs of DO terms with κ ≥ 0 or θ50 ≥ 0.2, from which a minimal spanning tree is constructed. (d,e) Zoomed-in views of selected disease labels around two network nodes.
