Figure 4.
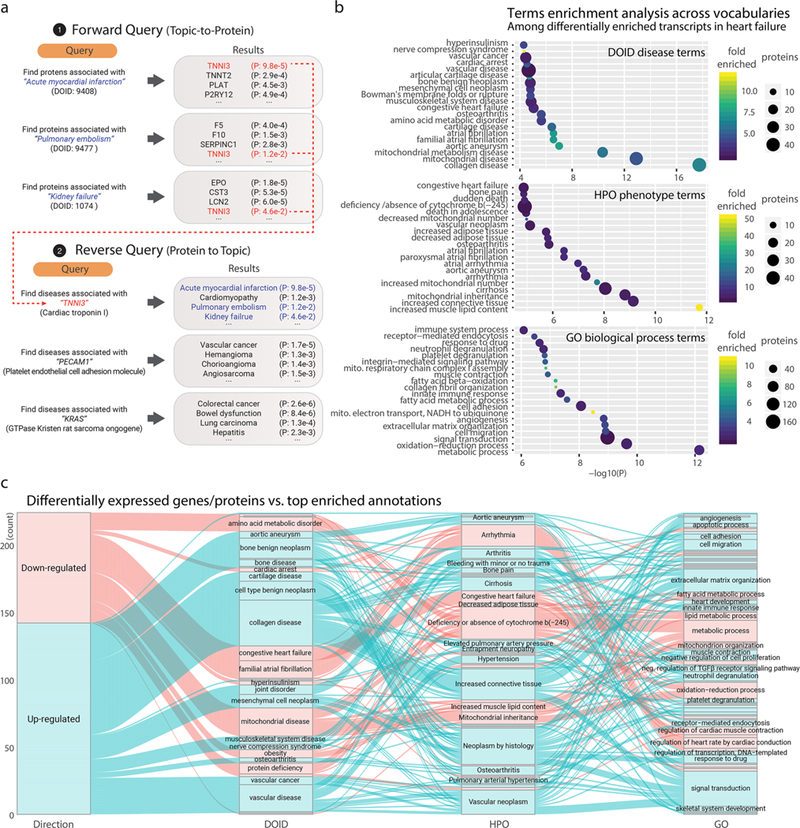
Enriched terms in reverse protein-to-disease query (DO and HPO) versus Gene Ontology. (a) Schematics for performing reverse (protein-to-term) queries using precompiled popular protein lists in the human diseasome. (b) Enriched terms (hypergeometric test P ≤ 0.05) from (top) DO, (middle) HPO, and (bottom) GO Biological Processes were associated with differentially expressed genes (limma adjusted, P ≤ 0.01) in a microarray data set from a rodent model of heart failure. (c) Relationship between assigned DO, HPO, and GO terms. Top associated terms are shown for each significantly up-regulated (blue) or down-regulated (red) transcript (limma adjusted P ≤ 0.01) in the microarray data set from a rodent model of heart failure. The alluvial streams link the top enriched term of DO to the corresponding terms in HPO and GO for each transcript. For example, a number of up-regulated transcripts are associated with the “familial atrial fibrillation” term in DO, corresponding in part to the “arrhythmia” term in HPO and to the “regulation of heart rate by cardiac conduction” term in GO.
