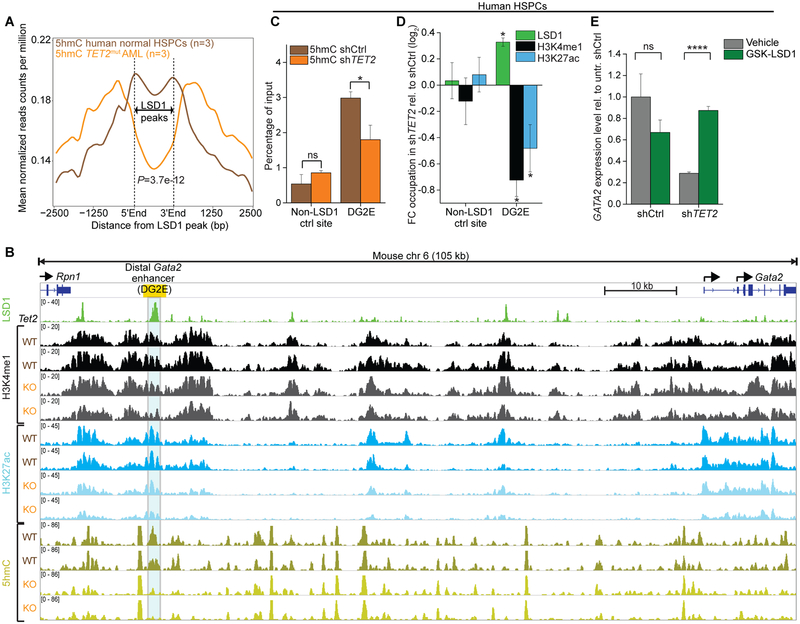
Figure 6.
Impairment of TET2 function facilitates LSD1-mediated enhancer inactivation. A, Averaged distribution of 5hmC profiles from hMe-Seal sequencing(11) at LSD1 binding peaks showed substantial reduction in primary human TET2mut AMLs (orange, n=3) in comparison to normal human CD34+ HSPCs (brown, n=3). P-value calculated by Wilcoxon-signed-rank test. B, IGV plot showing 5hmC DIP-seq and histone ChIP-seq tracks at the Gata2 locus (mm10, chr6:88,101,111–88,206,889) from in vitro cultured Tet2fl/fl pre-leukemic immature myeloid cells post-treatment with EtOH (WT, n=2) or 4-OHT (Cre-ERT2-mediated Tet2 deletion, KO, n=2)(14). LSD1 ChIP-seq(8) from murine immature myeloid 32D cells demonstrate strong enrichment of LSD1 at the distal Gata2 enhancer (DG2E; yellow box), where Tet2 deletion resulted in remarkable loss of 5hmC. C, QhMeDIP at the LSD1 target enhancer DG2E of GATA2 in human HSPC with TET2 knockdown or control shRNA. Bars represent mean enrichment over input with P-values compared to control (*P<0.05, Student’s t-test, means of triplicate measurements ± s.d). D, QChIP showing fold change of LSD1 binding as well as levels of H3K4me1 and H3K27ac after knockdown of TET2 relative to control in HSPCs (P<0.05, Student’s t-test compared to control shRNA, means of triplicate measurements ± s.d). E, Expression levels of GATA2 assessed by qPCR knockdown of TET2 or control in HSPCs (****P<0.0001, Student’s t-test, means of triplicate measurements ± s.d.). HSPCs were exposed to GSK-LSD1 treatment and measured at day 8 relative to untreated control shRNA set as 1 (normalized to COX6B1).