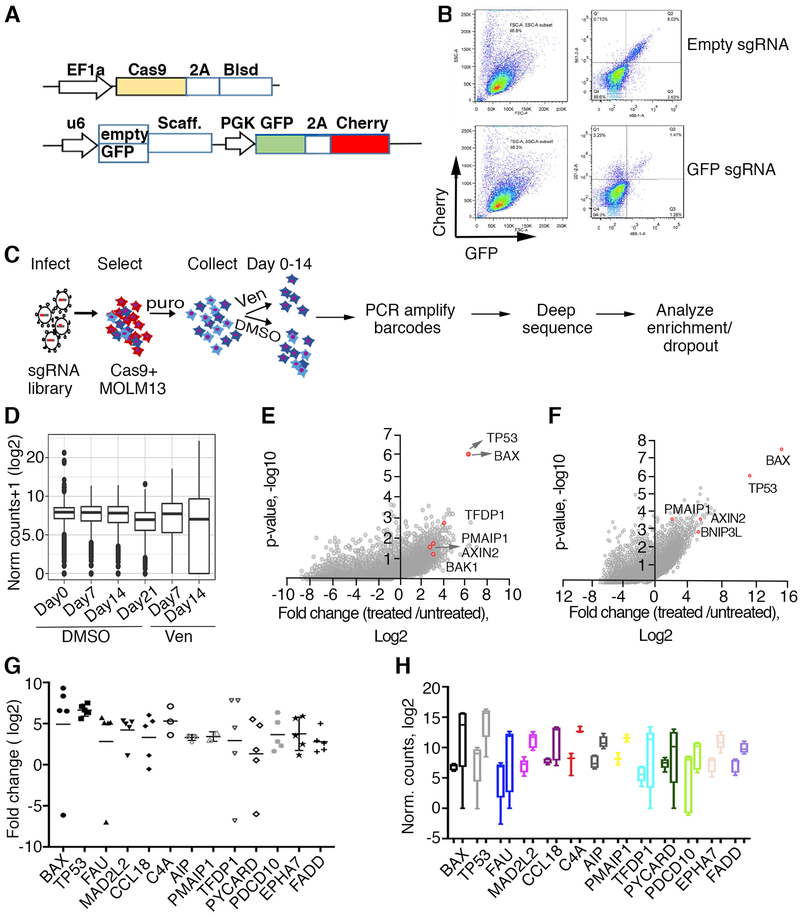
Figure 1. Genome-wide CRISPR/Cas9 screen in AML cells identified TP53, BAX and other apoptosis network genes conferring sensitivity to venetoclax.
A. Schematic representation of lentiviral vectors described elsewhere in detail [31] and used for delivery and functional assay of Cas9. Top: Cas-9 expressing vector; Blsd, blasticidin selection gene, EF1a, intron-containing human elongation factor 1a promoter, Cas9, codon-optimized Streptococcus pyogenes double-NLS-tagged Cas9, 2A, Thosea asigna virus 2A peptides. Bottom: vector carrying dual fluorescent proteins; GFP and mCherry expressed from the PGK promoter, U6 denotes human U6 promoter driving GFP sgRNAs or empty cassette, Scaff denotes sgRNA scaffold. B. Functional assay for Cas9 activity in MOLM-13 cells transduced with virus carrying an empty sgRNA cassette (top) or sgRNA targeting GFP (bottom), assessed by flow cytometry 5 days post transduction. Note the significant decrease in GFP signal in the presence of sgRNA targeting GFP. C. Schematic representation of genome wide screen for drug resistance. The sgRNA library [31] was transduced into Cas9-expressing MOLM-13 cells, selected with puromycin for the integration of sgRNA-carrying virus for 5 days and DNA collected from cells exposed to venetoclax (1 μM) or vehicle (DMSO) for various time points (days 0, 7, 14, 21). sgRNA barcodes were PCR-amplified and subjected to deep sequencing to analyze for enrichment and/or dropout. D. Normalized counts of sgRNAs from collected DNA samples, median, upper and lower quartiles are shown for representative replicate samples. E, F. Enrichment effect in Y. Kosuke (E) and Brunello (F) library screens for loss-of-sensitivity to venetoclax. Fold change and corresponding p-values are plotted; genes representing significant hits in both libraries are highlighted in red. G. Enrichment extent plotted as fold change over control following venetoclax exposure (day 14) for the set of individual top hit sgRNAs per gene is shown (Y. Kosuke library). H. Box and whisker plots spanning min/max values of normalized counts for control (left boxes in each pair) and venetoclax treatment (right boxes in each pair) combined for all sgRNAs per gene. Top hits are shown.