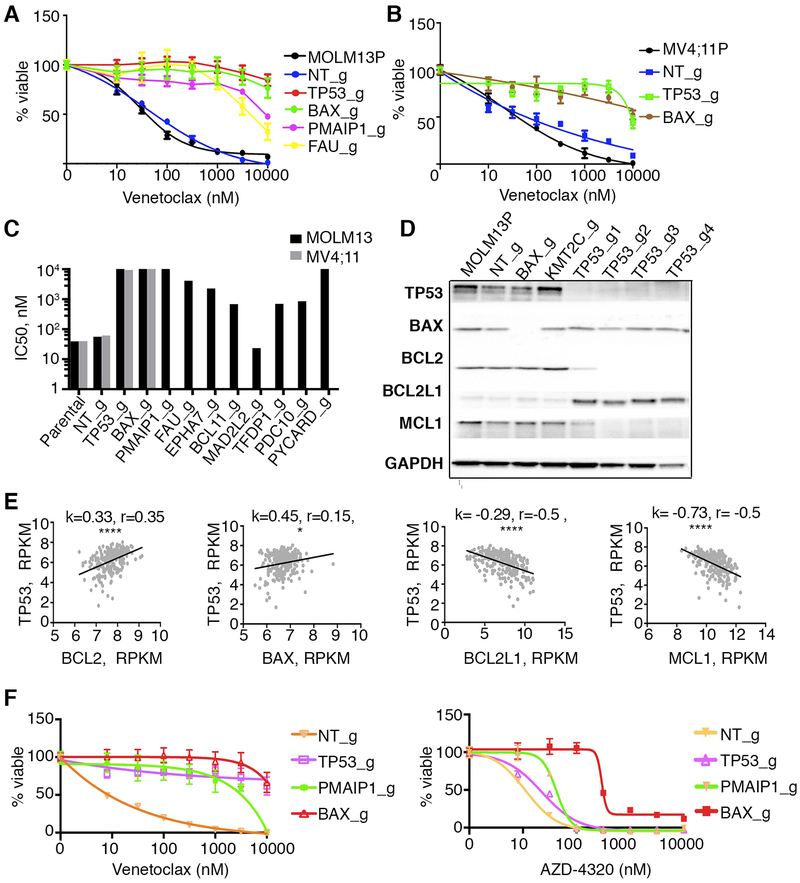
Figure 2. A. Confirmation of genes conferring venetoclax resistance in MOLM-13 and MV4;11 cells and correlations with changes in apoptotic gene transcription in AML patients.
MOLM-13 (A) or MV4;11 (B) cells were transduced with lentiviruses carrying single sgRNA/Cas9 constructs targeting TP53, BAX, PMAIP1, FAU or control (non-targeting; NT_g). 10 days post transduction, venetoclax sensitivity was measured in triplicate by colorimetric MTS assay using a 7-point concentration range (10 nM to 10 μM). Percentages of viable cells, after normalization to untreated controls, were fit using non-linear regression analyses; mean and standard errors are shown. MOLM-13P, parental MOLM-13 cells. MV4;11P, parental MV4;11 cells. C. Histogram (log scale) summarizing IC50 estimates from triplicate measurements for venetoclax sensitivity in parental MOLM-13 (black shaded), parental MV4;11 cells (grey shaded) or cells transduced with indicated sgRNA/Cas9 viruses. D. Western blot analyses of proteins extracted from MOLM-13 cells, transduced with indicated sgRNA/Cas9 viruses and identified with antisera to BAX, BCL2, BCL2L1(BCLXL), MCL1 and GAPDH. Note: for TP53, four sgRNAs producing distinct knockout alleles were used to confirm changes in expression levels of BCL2, BCL2L1(BCLXL), and MCL1. sgRNA targeting KMT2C was used as an additional unrelated sgRNA control. E. Correlation of expression of TP53 and selected genes in an AML patient sample cohort (n=246; [40]. k, slopes generated by linear regression; r, Spearman coefficient; **, p<0.01; ****, p<0.0001. F. Sensitivity profiles of venetoclax (left) and AZD4320 (right) on MOLM13 cells transduced with lentiviruses carrying single sgRNA/Cas9 constructs targeting TP53, BAX, PMAIP1, and NT (non-targeting) control, measured as in panel A using a 7-point concentration range 8 nM to 10 μM.