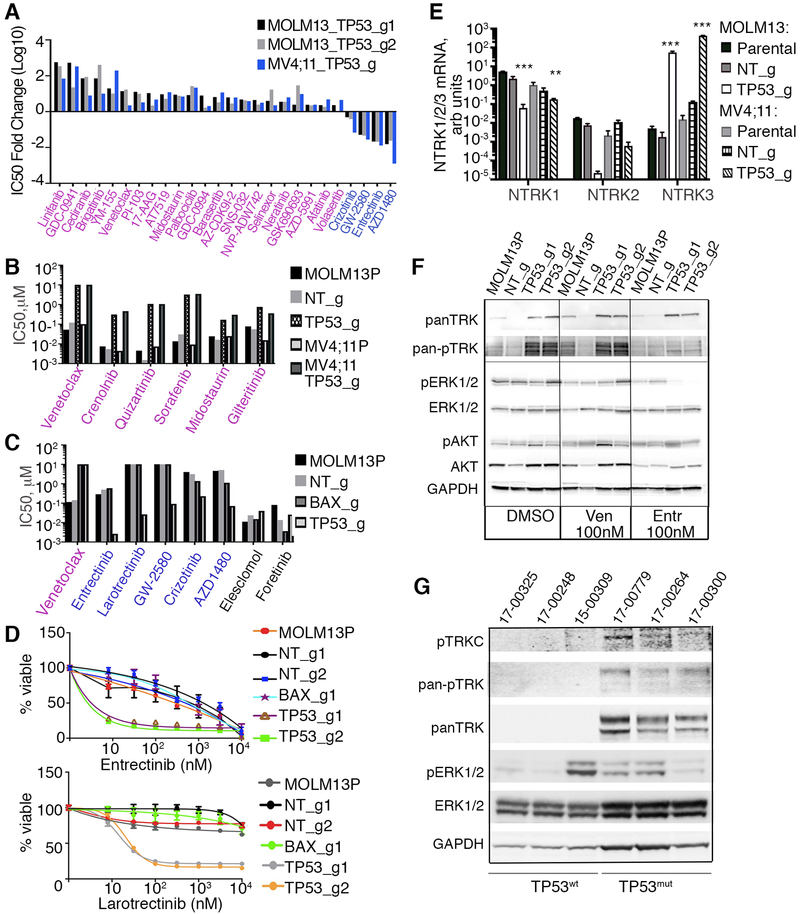
Figure 7. Cells with loss-of-function alleles for TP53 or BAX have altered sensitivities to small molecule inhibitors of various signaling pathways.
A. MOLM-13 and MV4;11 cells, transduced with indicated sgRNAs/Cas9 viruses, were screened with a panel of inhibitors targeting various molecular pathways and assessed using the drug screening method described in Figure 2A. Fold changes in IC50 values, (log10 scale) for those inhibitors concordant across both cells lines relative to nontargeting controls are shown. B. Analyses of FLT3 inhibitor sensitivities, in MOLM-13 and MV4;11 parental and KO cells, as indicated. Drug sensitivity was measured as described in Figure 2A, with IC50 estimated from triplicates. C. Histogram of sensitivities (IC50 values) to a series of NTRK inhibitors in MOLM-13 cells, transduced with indicated sgRNAs/Cas9 viruses. D. Drug sensitivity to NTRK inhibitors entrectinib (top) and larotrectinib (bottom) as measured in Figure 2A and 2F, in MOLM-13 cells transduced with indicated sgRNAs/Cas9 viruses. E. Expression levels of NTRK1, 2, and 3 mRNA using qRT-PCR analysis with RNAs isolated from MOLM-13 and MV4;11 cells with TP53 KO alleles or non-targeting (NT) control. Expression values were normalized to 36B4 gene expression levels. Statistical significance was determined ANOVA with Tukey posttest (**, p<0.01; ***, p<0.001). F. Western blot analyses of proteins extracted from MOLM-13 parental and MOLM-13 transduced with TP53 sgRNA/Cas9 viruses and treated overnight with venetoclax (Ven, 100 nM), entrectinib (Entr, 100 nM) or vehicle (DMSO), and identified with antisera to pan NTRK, phosphorylated NTRK (Tyr516), phosphorylated ERK1/2 (Thr202/Tyr204), ERK1/2, phosphorylated AKT(Thr308), AKT and GAPDH. G. Western blot analyses of proteins from AML patient samples with known TP53 mutant (TP53mut) or WT (TP53wt) status. Samples 17-00248 and 15-00309 have FLT3-ITD mutations; sample 17-00300 has a FLT3D835E mutation.