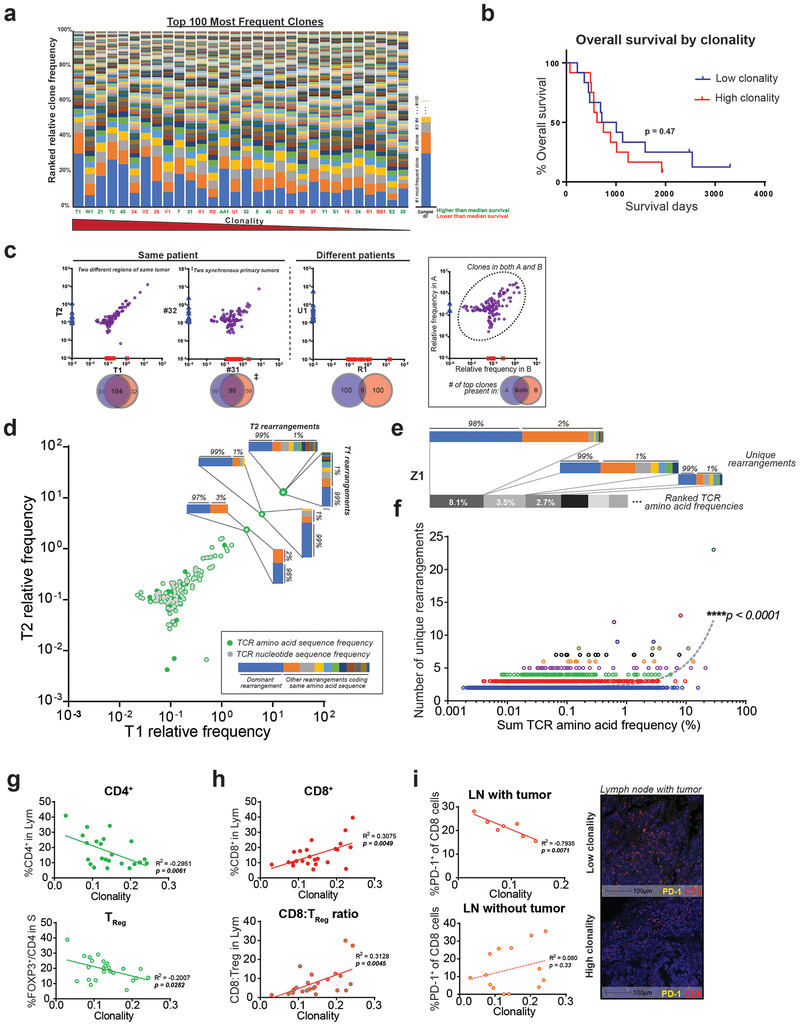
Figure 3: T cells clonally expand and demonstrate convergent evolution in the PDA tumor microenvironment.
(a) Clonality, as represented by the relative percentage of the 100 most frequent rearrangements within each sample. Arranged in decreasing clonality from left to right. Green sample ID refers to higher than median overall survival; orange is lower than median survival (n=30 samples). (b) Kaplan-Meier curve between low and high clonality patients, split by median (log-rank test, n=24). (c) Representative top 100 clone frequency plots of two samples from the same patient (n=6 pairs). Each axis represents relative frequency of clone in each sample; purple dots signify clones present in both samples. Venn diagram demonstrates the number of top 100 clones shared. (d-e) Representative plots showing multiple rearrangements code for the same amino acid TCR sequences. Each colored bar represents one unique DNA rearrangement. (f) Positive correlation between the relative frequency of an amino acid TCR sequence and the number of matched DNA rearrangements (linear regression, n=30). (g) Lower %TReg and (h) higher %CD8+ T cell correlates with higher clonality (linear regression, n=23–24). (i) PD-1 signal in tumor positive lymph node (LN) correlates negatively with clonality (representative multiplex image shown, scale bar is 100μm).