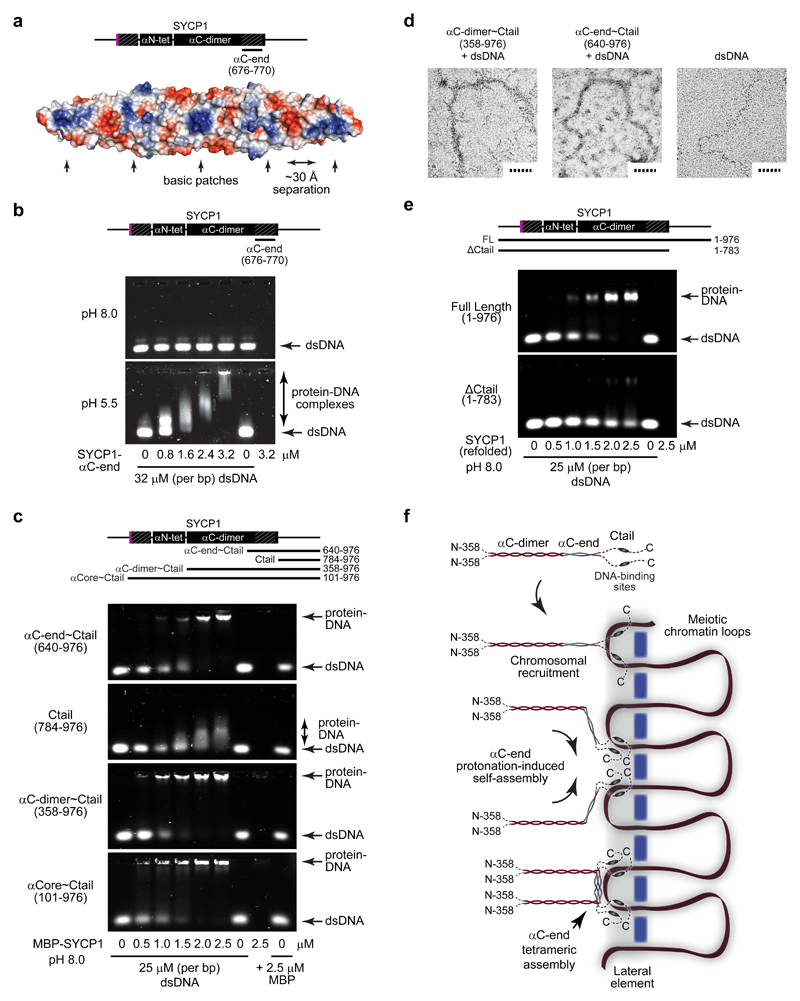
Figure 7. DNA binding by SYCP1.
(a) Surface electrostatic potential of the SYCP1 αC-end crystal structure (red – electronegative; blue – electropositive). The flat surface of the αC-end structure contains five demarcated basic patches that are separated by approximately 30 Å. (b) EMSA analysing the ability of SYCP1 αC-end (676-770) to interact with linear double-stranded DNA (dsDNA) at pH 8.0 (top) and pH 5.5 (bottom). Uncropped gel images are shown in Supplementary Data Set 1 (c) EMSA of MBP fusions of αC-end~Ctail (640-976), Ctail (784-976), αC-dimer~Ctail (358-976) and αCore~Ctail (101-976) with linear dsDNA at pH 8.0. (d) Electron microscopy (EM) analysis of MBP fusions of αC-dimer~Ctail (358-976) and αC-end~Ctail (640-976) in complex with plasmid dsDNA. Scale bars, 50 nm. (e) EMSA of refolded full length SYCP1 (1-976) and ΔCtail (1-783) with linear dsDNA at pH 8.0. (f) Model of SYCP1 chromosomal axis assembly. SYCP1 molecules are initially recruited to chromosomes through Ctail DNA-binding sites. The close proximity of DNA and/or interactions with chromosome axis proteins then triggers protonation-induced assembly of αC-ends into anti-parallel tetramers that bind DNA and thereby reinforce Ctail interactions. This results in the complete coating of the chromosome axis with SYCP1 molecules linked together through U-shaped assemblies that are anchored to chromosomal DNA.