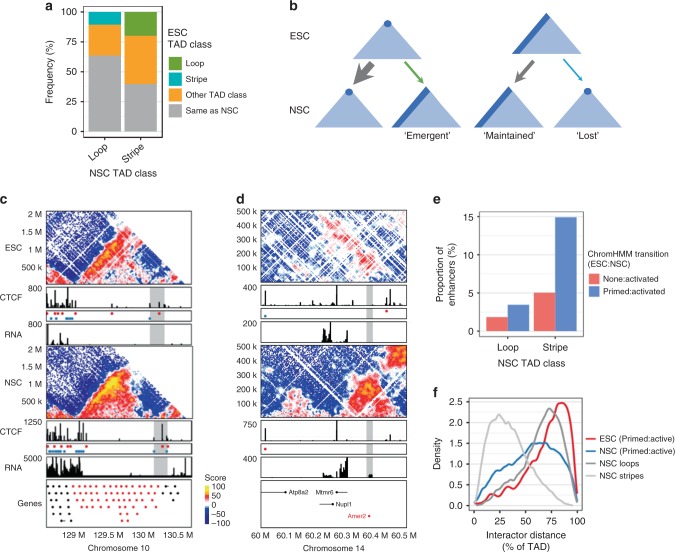
Fig. 5.
Structural changes to stripe TADs accompanying differentiation are coincident with changes in CTCF occupancy or lineage-appropriate enhancers. a TAD class dynamics between ESC and NSCs. ESC TADs separated into loop or stripe domains that maintain at least one border during differentiation, and their classification in NSCs. Frequency represents the proportion of NSC classified domains that were maintained or changed from a given ESC TAD group. b Schematic representation of data in (a) revealing the fates of TAD classes during differentiation. c, d Hi-C contact maps from ESCs and NSCs revealing TAD class changes associated with CTCF-binding site occupancy or transcription changes. CTCF ChIP-seq tracks, motifs, gene annotations, and gene expression are shown as before. c A 2.1 Mb region on chr10 around the Olf gene cluster changes from an ESC stripe to an NSC loop domain in conjunction with an increase in CTCF signal in NSCs at the unanchored stripe border (highlighted grey). d New insulation at a 500 kb region on chr14 is associated with a change from an ESC stripe to an NSC loop domain upon NSC-specific expression of Amer2. e Activated NSC enhancers defined by ChromHMM which were either unclassified (‘None’, red) or Primed (blue) in ESCs and their distribution in NSC loop or stripe domains. f Distribution of intra-TAD contact distance as a proportion of TAD size, in ESC (red) or NSC (blue) Hi-C maps from active NSC-specific enhancers which were primed in ESCs (Primed:active). As a comparison, the distribution of contact distances in loop (dark grey) or stripe (light grey) NSC domains are also shown. Note the shift between the blue and red distributions upon activation of NSC enhancers. Enhancer elements were expanded by 20 kb to include enough contact information