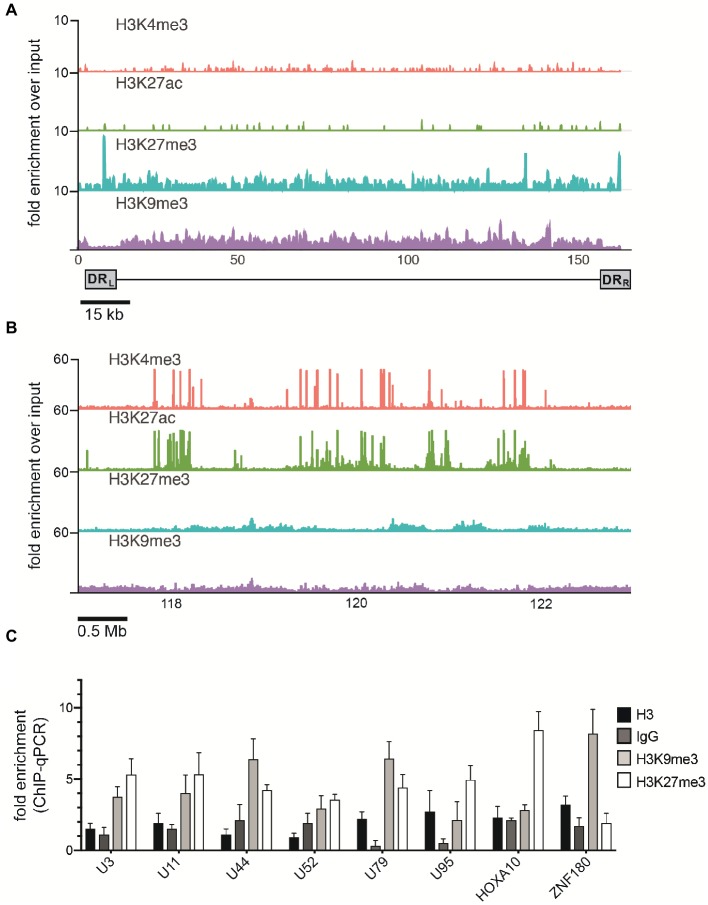
Figure 4.
ChIP enrichment profiles of histone modifications across the iciHHV-6A and human genomes. (A) The ChIP-seq enrichment patterns of two active (H3K27ac and H3K4me3) and two repressive modifications (H3K9me3 and H3K27me3) aligned to the HHV-6A (NC_001664.2) genome. The scale represents the fold enrichment of ChIP over input. (B) The ChIP-seq enrichment patterns of histone modification ChIP-seq datasets aligned to the human (hg38) genome. The ChIP-seq binding patterns demonstrate a typical profile of broad enrichment patterns of H3K27me3 and H3K9me3 in regions devoid of the active histone marks H3K27ac and H3K4me3. The locus at the indicated region of chromosome 2 of the hg38 human genome in mega base pairs is shown (Mb). (C) ChIP-qPCR assays using primers targeting HHV-6 gene regions U3, U11, U44, U52, U79, U95 and the cellular gene regions HOXA10 and ZNF180. Chromatin was immunoprecipitated with nonspecific IgG, total histone H3, H3K9me3, or H3K27me3 antibodies. Data are shown as the fold-enrichment over GAPDH normalized by input from three independent ChIP assays (mean ± SD).