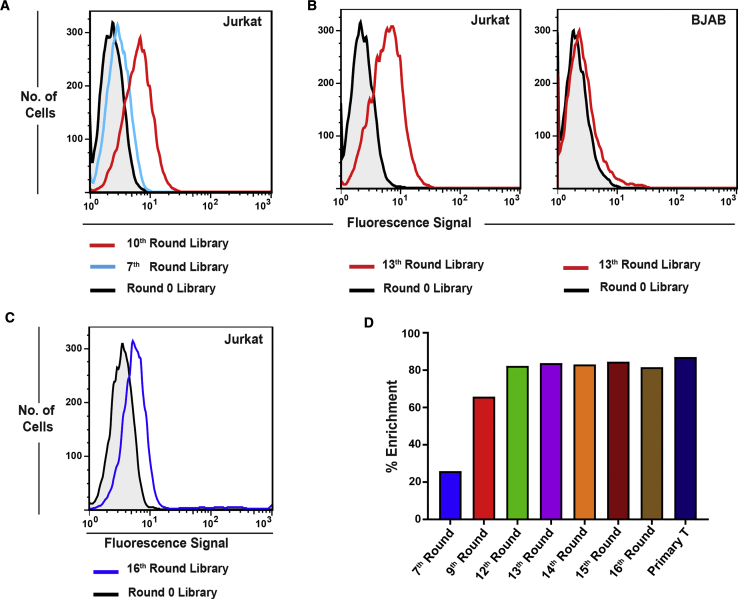
Figure 2.
Analyses of the Evolution of the SELEX Library against Jurkat Cells
(A) Flow-cytometric analysis of the evolved cell-SELEX library from 7th and 10th rounds against Jurkat cells. Higher fluorescence signal on the x axis indicates the higher binding of the fluorescence-labeled SELEX library from the 10th round against Jurkat cells compared to that of the fluorescence-labeled SELEX library from the 7th round and round 0. (B) Specificity analysis of the evolved cell-SELEX library by flow cytometry. The specificity of the 13th round of the cell-SELEX library was analyzed against Jurkat cells, using BJAB cells as the negative control. For the 13th round of the cell-SELEX library, the fluorescence signal on the x axis was higher against Jurkat cells, compared to that in round 0, but not against BJAB cells. This indicates that the cell-SELEX library evolved with sequences specific for Jurkat cells. (C) Flow cytometry analysis of binding of the 16th-round cell-SELEX library against a different batch of Jurkat cells. (D) Enrichment analysis of the sequencing data from cell-SELEX libraries based on FASTAptamer-Count results. Enrichment is defined as ; enrichment of the SELEX library increased as a function of cell-SELEX rounds until round 12, with no significant change in enrichment values beyond the 12th round.