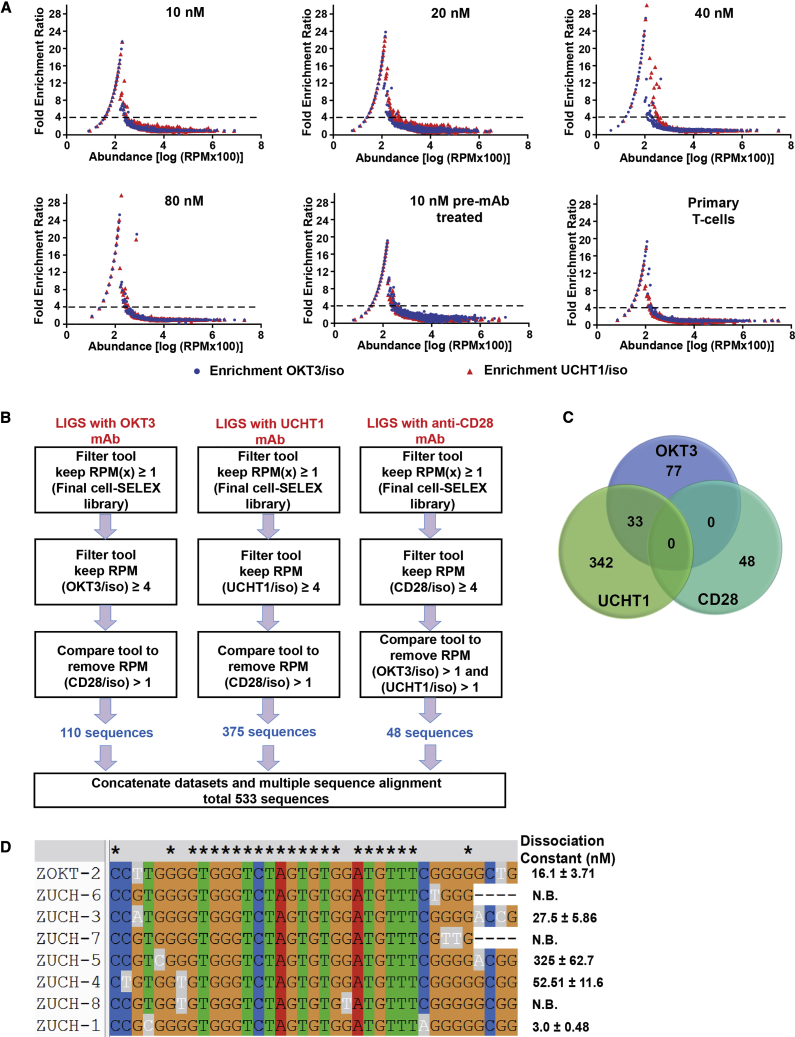
Figure 4.
Bioinformatics Analyses of Illumina HT Sequencing Data from LIGS Libraries
(A) The fold-enrichment ratios (RPMz/RPMy) against isotype control antibodies (y) and (z) represent either OKT3 (blue circles) or UCHT1 (red triangles) antibody, plotted as a function of the abundance of sequences. Here, the abundance values on the x axis are the log of normalized read counts (RPM) multiplied by 100., See Figure S4 for fold-enrichment ratios for anti-CD28. Based on the distribution of sequences, a fold-enrichment ratio value of 4 was chosen as a criterion for specificity (dashed line). (B) The schematic diagram summarizes downstream analysis using the GALAXY platform on FASTAptamer-Enrich data. LIGS libraries from the mAbs, including OKT3, UCHT1, and anti-CD28, were individually analyzed. After sequential filtering of sequences, 533 sequences in total were obtained for three mAbs used in LIGS. (C) Venn diagram summarizing the findings of downstream analyses using the GALAXY platform. Four hundred eighty-five sequences were identified in total by two different anti-CD3 antibodies, and 33 sequences were found to be common to both (OKT3 and UCHT1). (D) Multiple sequence alignment (CLUSTALW) of the identified aptamer family with their dissociation constants against Jurkat cells. The exceptional bases are highlighted in gray for each sequence. N.B., not binding. See Figure S7 for affinity curves for each aptamer.