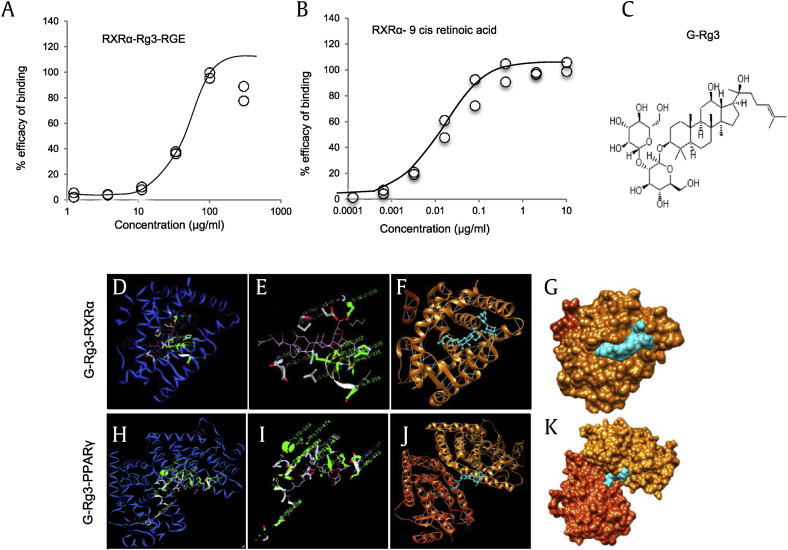
Fig. 1.
NHR-binding assay for Rg3-RGE with RXRα and 9-cis retinoic acid and molecular docking analysis of G-Rg3. The receptor-binding ligand assay was performed by Lead Hunter Discovery Services (Discover X Corporation). (A) Percent efficacy binding of RXRα-Rg3-RGE (Scatter graph details: Max = 100, Slope = 3.039, Min = 2.34, EC50 = 39.88, R2 = 0.966). (B) Percent efficacy binding of RXRα-9 cis retinoic acid (Scatter graph details: Max = 100.9, Slope = 0.9236, Min = 0.5399, EC50 = 0.01431, R2 = 0.9875). (C) Molecular structure of G-Rg3. (D–E) Solid surfaces showing the best docked complex with corresponding hydrogen binding sites (green). S-GLU-239 is the best docked position of RXRα, and S-LYS-319 is the best docked position of PPARγ (H–I) with the G-Rg3 ligand (central green spherule). (F and J) iGEMdock conformation cluster in a nonoverlapping manner with pockets (blue strings). White strings are indicative of hydrogen bonds and (G and K) 3D structures of the G-Rg3–RXRα complex and G-Rg3–PPARγ complex.3D, three-dimensional; G-Rg3, ginsenoside Rg3, NHR, nuclear hormone receptor; PPARγ, peroxisome-proliferating receptor γ; Rg3-RGE, Rg3-enriched red ginseng extract; RXRα, retinoid X receptor α.