Figure 2.
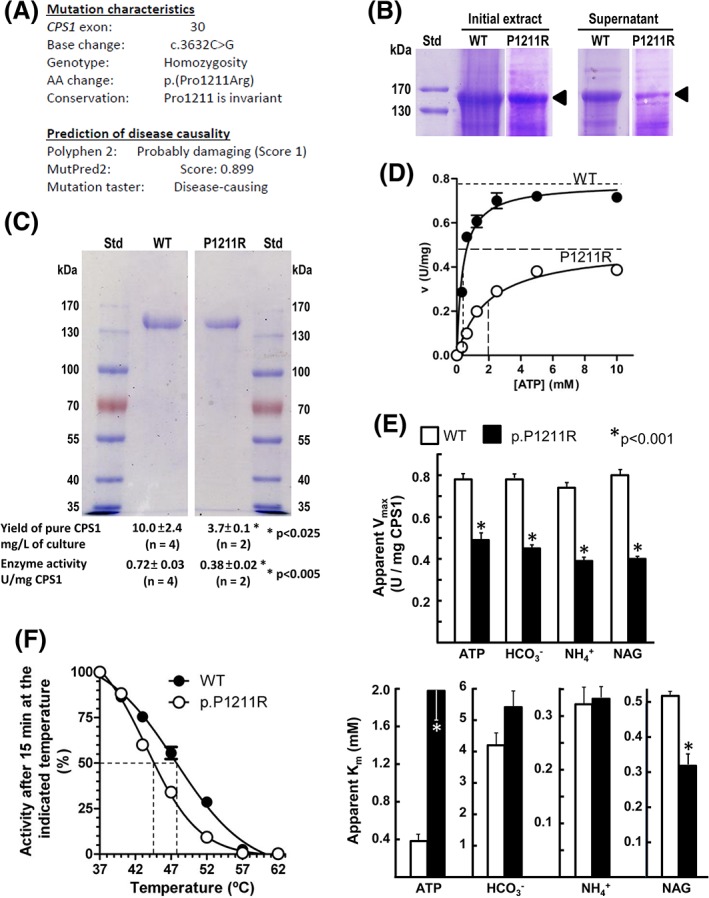
The p.(Pro1211Arg) mutation. A, Summary of the characteristics and in silico predictions for the mutation identified in our patient. (B) and (C) SDS‐PAGE of prestained protein standards (St) of the indicated masses (given on the side in kDa) and of wild‐type (WT) and p.(Pro1211Arg) (P1211R) CPS1 mutant of (B) total crude insect cell extracts and of the postcentrifugal supernatant, as indicated, to show the decreased solubility of the mutant form relative to WT CPS1 (CPS1 band marked with arrowpoint); or (C) after enzyme purification, with yields and enzyme activities of the purified proteins given below the tracks. Significant differences (Student t test) are given with an asterisk. D, Illustration of substrate kinetics for ATP for the wild type and the mutant form. Horizontal broken lines mark apparent V max values and vertical broken lines mark apparent K m ATP. E, Summary of V max (upper panel) and K m (lower panel) values for all three substrates. The asterisks denote significant differences for the indicated P value. F, Thermal inactivation experiments (for details see Section 2). Activities are expressed as a percentage of the activities observed after incubation at 37°C (at this temperature none of the two protein forms exhibited substantial activity losses). The broken vertical lines mark the temperatures of mid‐inactivation. CPS1, carbamoyl phosphate synthetase 1; NAG, N‐acetyl‐l‐glutamate
