Figure EV4. Yb bodies show characteristics of phase separation.
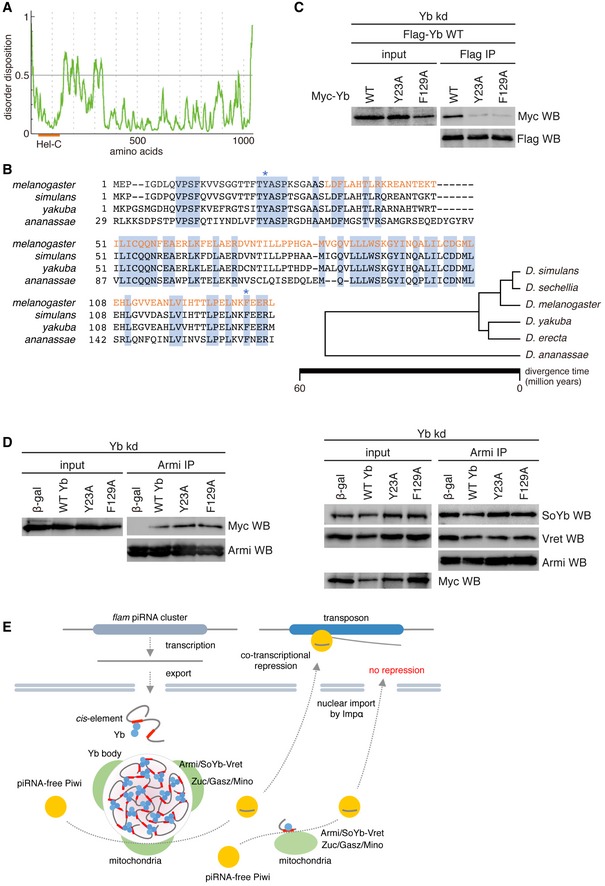
- PONDR‐FIT shows a potential IDR in Yb.
- Amino acid sequence alignment of the N‐terminal region of Yb in D. melanogaster (CG2076), D. simulans (JX64703.1), D. yakuba (XP_002100225), and D. ananassae (XP_001965435). The Hel‐C domain is indicated in orange. Amino acid residues conserved among four Drosophila members are highlighted in blue. Tyr23 and Phe129 in Yb in D. melanogaster are shown by blue asterisks. The family tree of flies in the D. melanogaster group was adapted from Ref. 61.
- Yb Y23A and F129A mutants barely interacted with WT Yb.
- Yb Y23A and F129A mutants associated with Armi (left). Formation of Armi/SoYb/Vret complex was hardly affected by the mutants (right).
- Proposed model for Yb body formation and piRNA biogenesis in OSCs. Homotypic Yb protein (light blue on left) binds flam RNA transcripts through their Yb binding sites (red bars). The Yb–Yb association and Yb–RNA interaction lead to the assembly of multivalent Yb bodies. Yb is shown in a trimer form for simplification, although it may multimerize in vivo. Yb bodies are often surrounded by mitochondria, on the surface of which piRNA biogenesis factors Zuc, Gasz, and Mino are present. At the interface of the two organelles (piRNA biogenesis site), flam‐piRNA biogenesis occurs. Piwi bound to flam‐piRNAs (Piwi‐piRISC) is localized to the nucleus and silences transposons cotranscriptionally. Some piRNAs are produced independent of the Yb bodies. This happens when Yb lacks the Hel‐C domain, leading to the failure of multivalent Yb body formation. Genic piRNAs may be produced in this way even in normal OSCs. Flam‐piRNAs repress transposons but genic piRNAs are theoretically unable to target transposons. Piwi bound to genic piRNAs can also be localized to the nucleus, but is useless in transposon repression.
Source data are available online for this figure.
