Figure 1. Characteristics of NSL complex proteins.
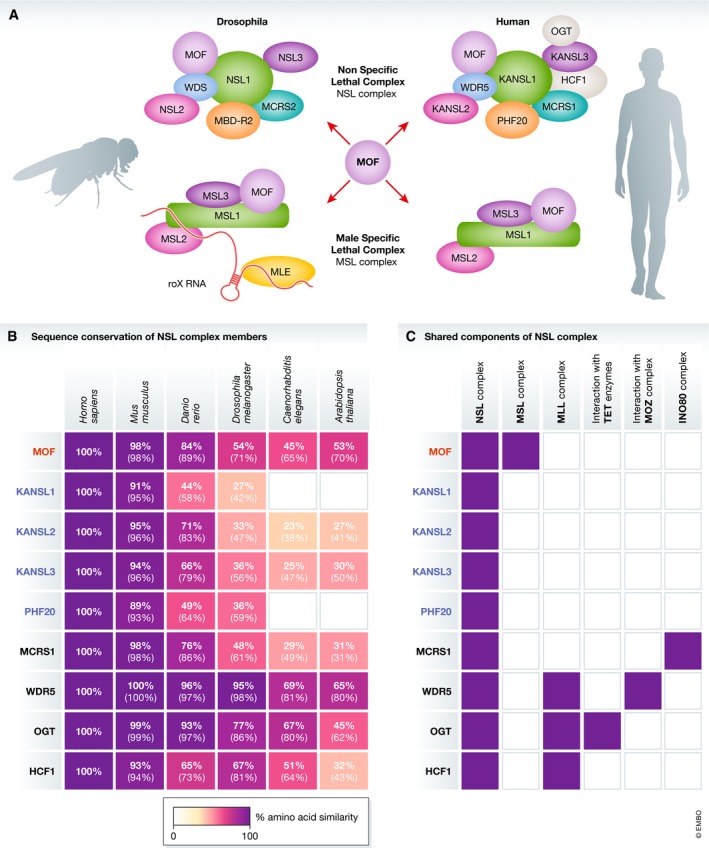
(A) The two MOF‐containing complexes—the NSL and MSL complexes. The NSL and MSL complex components in the two best studied models, D. melanogaster and human, are depicted. (B) Conservation of the NSL complex through evolution. The heatmap indicates the level of amino acid similarity in the conserved domains of NSL complex members. The percentages written over the heatmap indicate the precise amino acid identity in the conserved domains, with the percentage similarity is provided in the parenthesis. The human NSL protein information was extracted from the NCBI protein database followed by blast of the whole protein sequence to determine the identity of NSL complex orthologs and amino acid conservation. (C) Core versus shared members of the NSL complex. MOF, the catalytic subunit of the NSL complex, is also found in the MSL complex 40. OGT also interacts with the TET enzymes TET1‐3 that modify the DNA 5‐methyl‐cytosine base to 5‐hydroxymethyl‐cytosine 104, 105, 106, 107, as well as the MLL/SET complexes 108. In addition to the NSL complex, WDR5 also associates with the MLL trithorax 109 and the MOZ acetyltransferase 110 complexes. HCF1 is shared with the MLL trithorax complex 108, 109, 111, while MCRS1 co‐immunoprecipitates both NSL and INO80 complex members 40. In contrast, KANSL1, KANSL2, KANSL3 and PHF20 have only been identified as part of the NSL complex to date.
