Figure 3. Multiple facets of the NSL complex.
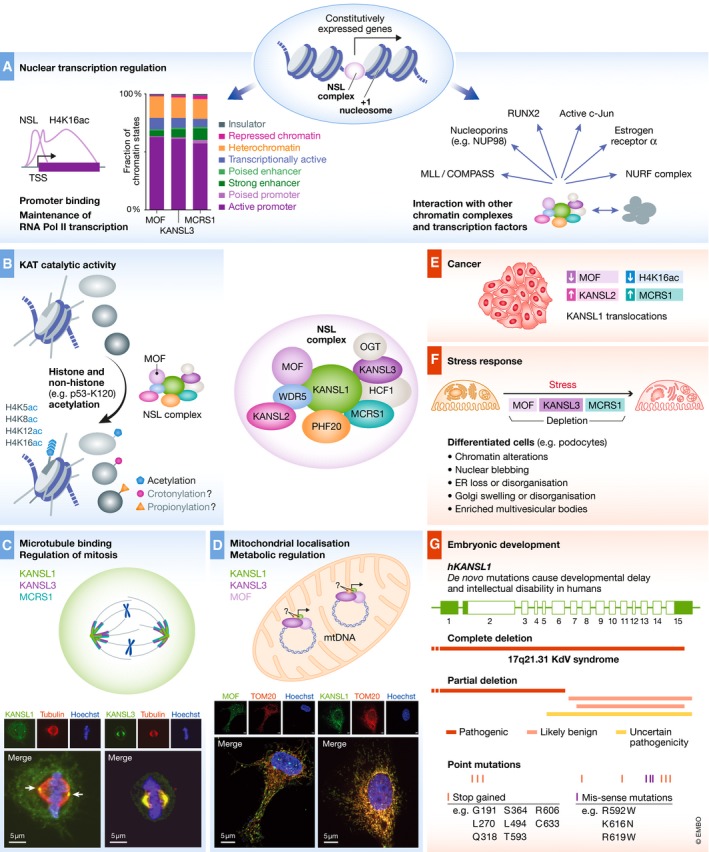
(A) The NSL complex regulates nuclear gene transcription. In D. melanogaster, the NSL complex localizes to gene promoters and maintains the positioning of the +1 nucleosome (middle panel). In addition, the NSL complex acetylates H4K16, which is enriched at promoters as well as gene bodies of transcriptionally active genes (left panel). A similar chromatin‐binding pattern is observed in mammalian cells, whereby MOF, KANSL3, and MCRS1 localize to active gene promoters and to a lesser extent, to enhancers in mouse ESCs. The plot on the left side of panel (A) was generated by apportioning all NSL‐bound regions in ChIP‐seq data from male mouse ES cells 35 into different chromatin states 114. To mediate transcriptional control, the MOF‐NSL complex interacts with other chromatin complexes and transcription factors, allowing for crosstalk and collaboration between different signaling pathways and chromatin regulators (right panel). (B) Besides histones, MOF acetylates non‐histone proteins such as p53 115. This encourages investigation of MOF partner proteins and MOF targets outside of chromatin. Moreover, MOF has been implicated in catalyzing other –acyl chains such as crotonylation and propionylation 116, 117. However, the precise contribution of MOF and its NSL complex members to non‐acetyl acylations needs further investigation. (C) KANSL1, KANSL3, and MCRS1 localize to microtubules during mitosis and are required for the progression of cells through cell division. (D) The NSL complex members MOF, KANSL1, and KANSL3 localize to the mitochondria. This finding has opened up a new horizon for the study of the predominantly chromatin‐associated NSL complex in the regulation of mitochondrial biology. (E) MOF, KANSL1, KANSL2, and MCRS1 are dysregulated in various types of cancer, and the misregulation of NSL complex members drives tumor pathology and aggressiveness. (F) The MOF‐NSL complex is implicated in multi‐organellar stress responses by maintaining key transcriptional networks such as autophagy and DNA repair. (G) Heterozygous deletion or point mutations in human KANSL1 (adapted from patient entries on DECIPHER GRCh37) cause the KdV syndrome, which is typified by developmental abnormalities and intellectual disability. How the NSL complex regulates development and brain function, however, is an outstanding question.
