Abstract
Epithelial ovarian cancer (EOC) is one of the most common malignant gynecological tumors. Interval cytoreductive surgery and cisplatin-based chemotherapy are the standard treatments. However, acquired resistance to cisplatin presents a major challenge for improving the overall survival and prognosis of patients. Recent evidence indicates that cytoprotective functions of autophagy in cancer cells is a potential mechanism for chemoresistance. The present study aimed to investigate the mechanisms responsible for cisplatin resistance in EOC cell lines. The results revealed that cisplatin activated autophagy, measured by an increase in the expression of LC3-II by western blot analysis, protecting ovarian cancer cells from cisplatin toxicity. The present study also identified Bcl-2-associated athanogene 3 (BAG3) as a novel autophagy regulator that serves a role in cisplatin resistance. Treatment with cisplatin was observed to enhance BAG3 expression in parental and cisplatin-resistant ovarian cancer cell lines, and the downregulation of BAG3 blocked cisplatin-induced autophagy, thereby increasing cisplatin sensitivity in the EOC cell lines. In conclusion, BAG3 attenuates cisplatin resistance by inhibiting autophagy, suggesting that downregulation of BAG3 may be a useful therapeutic strategy to overcome cisplatin resistance by preventing cytoprotective autophagy in EOC.
Keywords: BAG3, autophagy, ovarian cancer, cisplatin, chemoresistance
Introduction
Epithelial ovarian cancer (EOC) accounted for ~90% of ovarian cancer cases worldwide in 2012 (1). EOC remains the most lethal gynecological malignancy due to the heterogeneous nature of the disease, and the majority of cases are diagnosed at a late stage (2). The principal treatments for ovarian cancer are complete surgical staging and maximal resection with cisplatin-based chemotherapy (3). However, chemoresistance against platinum-based therapies can decrease chemotherapeutic efficacy and culminate in tumor recurrence (4). Therefore, it is imperative to explore the mechanisms of chemoresistance and to develop effective therapeutics for the treatment of ovarian cancer.
Cisplatin was introduced into clinical trials in 1971, and is widely used in the treatment of solid tumors, such as ovarian cancer (5). This compound is the core drug used in combination with other chemotherapeutic agents in the treatment of ovarian cancer in the clinical setting (6). However, primary or acquired drug resistance is a major challenge and decreases the treatment efficiency (7). The potential mechanisms underlying platinum-based chemotherapy involve pharmacological, biochemical, apoptosis and microenvironment resistance (8). Emerging evidences have also indicated that autophagy may be involved in cell survival and chemoresistance mechanisms (9–12). Macroautophagy (autophagy) is a conserved catabolic pathway in which cytosolic contents, such as damaged organelles, misfolded proteins and bacteria, are transported into lysosomes for degradation (13,14). Cytotoxic drugs often trigger autophagy, particularly in apoptosis-defective cells (15); however, how the tumor cells utilize autophagy to prevent chemotherapy-mediated cell death remains elusive.
The Bcl-2-associated athanogene 3 (BAG3) protein is a member of the BAG family (16). The human BAG3 protein contains a C-terminal heat shock protein 70-binding BAG domain, which inhibits its chaperone activity (17), a WW domain, a proline-rich (PXXP) domain, and two conserved IPV motifs, one located centrally and the other closer to the N terminus (18). BAG3 not only interacts with Bcl-2 to inhibit apoptosis (16,19), but also participates in the regulation of autophagy by interacting with heat shock protein family proteins and heat shock protein β-8, to facilitate the removal of misfolded and degraded proteins (20). The human BAG3 gene is located on the long arm of chromosome 10 26.11 and is expressed constitutively in myocytes, certain other normal cell types, such as breast and thyroid cell lines, and several primary tumors or tumor cell lines, including ovarian cancer cells (21). Numerous types of cell factors can induce the expression of BAG3. For example, stress, heavy metals and HIV infection can upregulate BAG3 expression (22). BAG3 dysfunction has been implicated in numerous disorders, including cancer, myopathies and neurodegeneration. Previous evidence has suggested that the BAG3 protein exerts a function in regulating the balance between cell survival and death (16). A previous study reported that BAG3-mediated selective macroautophagy is an adaptive mechanism that maintains cellular homeostasis under stress conditions and during aging, making BAG3 a potential target for future pharmacological interventions (16). However, whether BAG3 participates in cisplatin resistance in ovarian cancer cells, and the underlying mechanisms remain unclear.
The present study utilized the cisplatin-resistant ovarian cancer cell line SKOV3/DDP and the parental SKOV3 cell line to identify: i) The roles of BAG3 in cisplatin-induced autophagy associated with the drug resistance and ii) the relevant mechanisms for regulating the process. The present findings support BAG3 as a potential therapeutic target for enhancing the efficacy of cisplatin in ovarian cancer.
Materials and methods
Cell lines and reagents
The human EOC cell lines SKOV3 (parental) and SKOV3/DDP (cisplatin-resistant) were obtained from the Cell Bank of the Chinese Academy of Sciences (Beijing, China). The cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 100 U/ml streptomycin/penicillin at 37°C in a humidified atmosphere containing 95% air and 5% CO2. The SKOV3/DDP cells were additionally cultured with 1 µM cisplatin to maintain drug-resistance. Antibodies against microtubule-associated protein 1 light chain 3 (LC3; cat. no. 2775), sequestosome 1 (p62/SQSTM1; cat. no. 5114), caspase-3 (cat. no. 9662), cleaved caspase-3 (cat. no. 9664), poly [ADP-ribose] polymerase (PARP; cat. no. 6542) and cleaved PARP (cat. no. 5625) were obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA). Antibodies against GAPDH (cat. no. 10494-1-AP) and BAG3 (cat. no. 10599-1-AP) were purchased from Protein Technology (Chicago, IL, USA). Cisplatin (cat. no. BP809, 15663-27-1), chloroquine disphosphate (CQ; cat. no. C6628, 50-63-5) and the secondary antibody (anti-mouse immunoglobulin G; cat. no. A2304) were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). The cell culture medium was purchased from Gibco (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cell viability assay
SKOV3 (1×104) and SKOV3/DDP (6×103) cells were seeded into 96-well plates and cultured in the aforementioned conditions for 24 h. Cells were permitted to adhere, and were then pretreated at 37°C with or without 10 µM CQ for 2 h, followed by the indicated concentrations of cisplatin (20 µM), CQ (10 µM) or a combination (20 µM cisplatin + 10 µM CQ) for the indicated times. DMSO was used as the untreated control. In brief, 10 µl Cell Counting Kit-8 (CCK8) reagents (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) were added to each well and cultured at 37°C for ~1 h, and the absorbance at 450 nm was then calculated using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Cell survival was calculated as: Cell survival rate (%)=(experimental group OD value-blank group OD value)/(control group OD value-blank group OD value). The concentration of cisplatin necessary to cause 50% inhibition (IC50) was calculated by non-linear regression using GraphPad Prism version 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). All samples were performed in triplicate for each group.
Green fluorescent protein (GFP)-LC3 plasmid transfection and autophagic flux assay
The SKOV3 and SKOV3/DDP cells were transfected with a GFP-LC3 expression plasmid (500 ng/µl, BioVector, Ltd, Preston, UK) at 70–80% confluence using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). The time interval between transfection and subsequent experimentation was 48 h. After 48 h, the cells were fixed with 3.7% formaldehyde for 15 min at room temperature and washed three times with cold PBS, and the distribution and fluorescence of GFP-LC3 were visualized by fluorescence microscopy (magnification, ×200). The numbers of GFP-LC3 puncta were counted manually in 20 cells per group, in 10 randomly selected fields of view as described previously (23).
Transmission electron microscopy (TEM) assay
Following treatment, cells were washed with 0.1× cacodylate buffer (pH 7.4) and were fixed with 2% glutaraldehyde in PBS for 24 h at 4°C. The rest of the process was conducted as described previously (24). Sections were examined using a Zeiss transmission electron microscope (magnification, ×10,000; Zeiss GmbH, Jena, Germany).
Flow cytometry assay
The degree of apoptosis was assessed using an Annexin V-FITC/propidium iodide (PI) apoptosis detection kit (BD Pharmingen; BD Biosciences, San Jose, CA, USA). Following the aforementioned treatments, cells were harvested in 0.25% trypsin and washed three times with PBS, and 1×105 cells were then suspended in 500 µl binding buffer and incubated with 5 µl Annexin V and 5 µl PI for 15 min in the dark. Analysis of apoptotic cells was performed using a flow cytometer (Cytomics™ FC 500; Beckman Coulter, Inc., Brea, CA, USA; CXP. Software version 1.0 CXP Cytometer; Beckman Coulter, Inc.).
TUNEL assay
A total of 3×105 SKOV3 and SKOV3/DDP cells per well were seeded on six-well plates and fixed with 3.7% paraformaldehyde at room temperature for 1 h. The cells were permitted to adhere. The cells were washed three times with PBS for 5 min between each step, which was followed by the addition of 100 µl 0.1% Triton X-100. The cells were then placed on ice for 2 min and 50 µl TUNEL (Beyotime Institute of Biotechnology, Shanghai, China) reaction mixture (2 µl TdT enzyme and 45 µl fluorescence labeling) was added to the cells, followed by incubation at 37°C for 1 h. Cell nuclei were stained with 1 ml 10 µg/ml DAPI (Beyotime Institute of Biotechnology) for 5 min at room temperature in the dark and observed under a fluorescence microscope for TUNEL and DAPI staining (EVOS™ Thermo Fisher Scientific, Inc.). Cells with red fluorescence were defined as apoptotic cells, and the number of apoptotic cells was counted in 5 randomly selected fields of view at ×200 magnification.
cDNA constructs, short hairpin (sh)RNA and transfection
Cells were transfected with a GFP-tagged LC3 cDNA expression construct or BAG3 shRNA (GenePharma, Shanghai, China) using Lipofectamine 3000, according to the manufacturer's protocol. The sequence of BAG3 shRNA was 5′-GATACACGAGCAGAACGTTAC-3′, the concentration of BAG3 shRNA used for transfection was 500 ng/µl. The time interval between transfection and subsequent experimentation was 48 h.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from SKOV3 and SKOV3/DDP cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and RT was performed using the PrimeScript™ RT Master Mix (Takara Bio, Inc., Shiga, Japan). The temperature protocol used for RT was, 37°C for 15 min, 85°C for 5 sec followed by a 4°C holding step. The qPCR reactions were performed using SYBR Green reagents (Takara Bio Inc.) on an Agilent Mx3005P system (Agilent Technologies, Inc., Santa Clara, CA, USA). The thermocycling conditions were: 95°C for 30 sec; 35 cycles of 95°C for 30 sec; 60°C for 30 sec; 72°C for 15 sec, fluorescent signals are collected here; 95°C for 5 sec; and 65°C for 59 sec; and 95°C for 30 sec. Each sample was run in triplicate. The primers used were as follows: Human BAG3 forward, 5′-CTCCATTCCGGTGATACACGA-3′ and reverse, 5′-TGGTGGGTCTGGTACTCCC-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′. The relative expression levels were calculated using the 2−ΔΔCq method (25).
Immunofluorescence assay
SKOV3 and SKOV3/DDP cells were cultured on coverslips, fixed with 3.7% paraformaldehyde in PBS for 30 min at room temperature, and permeabilized with 0.1% Triton X-100 in PBS for 20 min. Following blocking with 10% FBS for 30 min at room temperature, cells were incubated with primary antibodies for 2 h, washed three times with PBS, and incubated with secondary antibodies for 1 h at room temperature in the dark. The nuclei were then visualized using DAPI for 5 min at room temperature in the dark (Beyotime Institute of Biotechnology) and the cells were observed using a Leica TCS SP5 laser confocal microscope (magnification, ×800; Leica Microsystems GmbH, Wetzlar, Germany).
Western blot analysis
Following the aforementioned treatments, SKOV3 and SKOV3/DDP cells were washed with ice-cold PBS and lysed in RIPA buffer (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China), and protein concentrations were subsequently determined by a bicinchoninic acid protein assay (Beyotime Biotechnology Institute of Biotechnology). The protein lysates (40 µg) were separated by 12% SDS-PAGE or 15% SDS-PAGE and transferred to PVDF membranes (EMD Millipore Corp., Billerica, MA, USA). The membranes were blocked with 5% skimmed milk for 2 h at room temperature with slow agitation, incubated with primary antibodies (1:1,000) overnight at 4°C, and were subsequently incubated with horseradish peroxidase-conjugated secondary antibodies (1:2,000) for 1 h at room temperature. The membranes were then visualized with ECL reagents (Applygen, Beijing, China). Densitometric analysis was performed using the ImageJ software version 6.0 (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
All the data are presented as the mean ± standard deviation. Data were analyzed and visualized using GraphPad Prism version 5.0. Two-sided Student's t-test was used to assess for statistically significant differences between two groups. A one-way analysis of variance followed by Tukey's post-hoc test was used to determine statistical differences among multiple treatment groups. All experiments were independently repeated in triplicate. P<0.05 was considered to indicate a statistically significant difference; all tests were two-sided and no corrections were applied for multiple significance testing.
Results
Sensitivity of EOC cell lines to cisplatin treatment
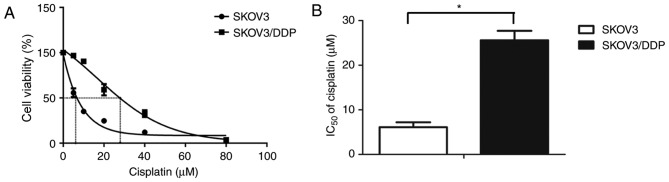
The present study detected cisplatin sensitivity of the SKOV3/DDP cells compared with their parental SKOV3 cells using the CCK8 assay after the cells were exposed to various concentrations of cisplatin for 48 h. The percentage of surviving cells decreased in a dose-dependent manner in the SKOV3 and SKOV3/DDP cells (Fig. 1A). The 48 h IC50 values of cisplatin in SKOV3 and SKOV3/DDP cells were 6.12±1.09 and 25.59±2.116 µM, respectively (Fig. 1B). Based on these results, 20 µM cisplatin was selected for use in the subsequent experiments.
Figure 1.
Sensitivity of epithelial ovarian cancer cells to cisplatin treatment. (A) SKOV3 and SKOV3/DDP cells were treated with various concentrations of cisplatin for 48 h, and the cell viability was determined using a Cell Counting Kit-8. The dose response curves presented as a sigmoidal shape. (B) The IC50 values of cisplatin in SKOV3 and SKOV3/DDP cells were calculated. Each value represents the mean ± standard deviation from three independent experiments. *P<0.05. IC50, half maximal inhibitory concentration.
Cisplatin induces cytoprotective autophagy in EOC cells
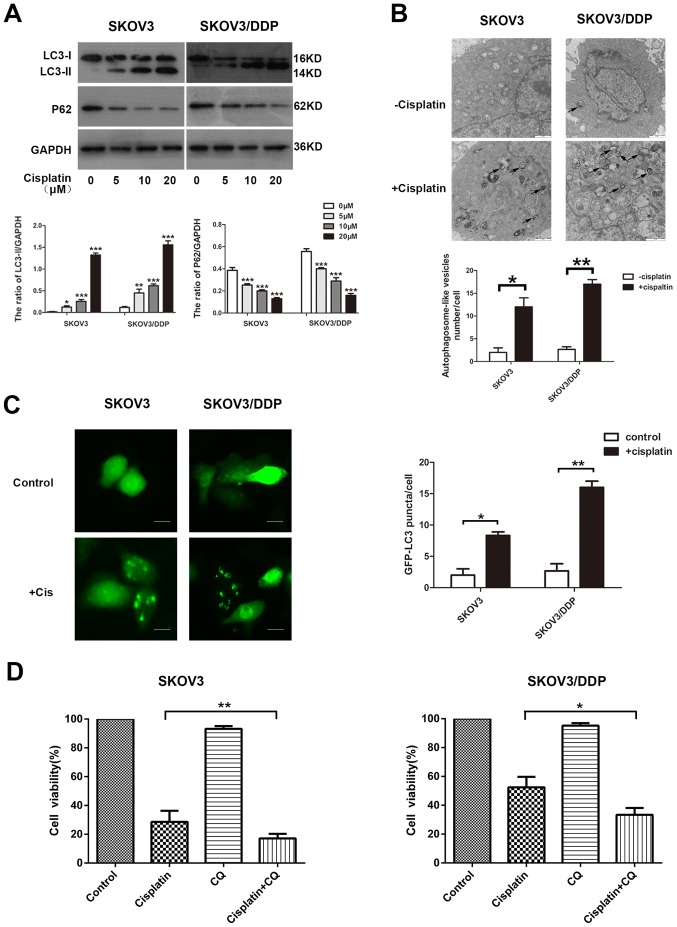
The ability of cisplatin to induce autophagy in EOC cells was investigated. The detection of LC3, a mammalian ortholog of yeast Atg8, is a common way to monitor autophagy. During autophagy induction, the non-lipidated form of LC3 (LC3-I) is conjugated with phosphatidylethanolamine, converted into the lipidated form (LC3-II), which is associated with autophagosome biogenesis. The p62 protein links with LC3-II or ubiquitinated substrates, which are degraded during autophagic flux (26). In the present study, SKOV3 and SOV3/DPP cells were exposed to concentrations of cisplatin ranging from 0 to 20 µM, and the drug was observed to activate autophagy. The treatment increased the level of LC3-II and decreased p62 expression in the SKOV3 and SKOV3/DDP cells, as demonstrated by western blotting (Fig. 2A). Similar results were observed with ultrastructural analysis of autophagosomes using TEM (Fig. 2B). The effect of cisplatin on autophagy was further confirmed by a GFP-LC3 punctate-formation assay. SKOV3 and SKOV3/DDP cells were transiently transfected with a GFP-LC3 plasmid to monitor autophagosome formation through fluorescence microscopy, and the number of GFP-LC3 puncta cells were increased in the examined cell lines (Fig. 2C).
Figure 2.
Cisplatin induces cytoprotective autophagy in epithelial ovarian cancer cells. (A) Parental SKOV3 and cisplatin-resistant SKOV3/DDP cells were treated with the indicated concentrations of cisplatin for 48 h and whole-cell lysates were subjected to western blot analysis to detect the protein levels of LC3-I, LC3-II and p62. GAPDH served as a loading control. Densitometric values were quantified and the data are presented as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. the control group. (B) Autophagic vacuoles were detected by transmission electron microscopy. The black arrows indicate autophagic vacuoles. Scale bar, 1 µm. *P<0.05 and **P<0.01. (C) SKOV3 and SKOV3/DDP cells were transiently transfected with a GFP-LC3 expression plasmid for 48 h and then exposed to cisplatin (20 µM) for an additional 48 h. The formation of GFP-LC3 puncta was examined using immunofluorescence and quantified. Each value represents the mean ± standard deviation from three independent experiments. Scale bar, 200 µm. *P<0.05 and **P<0.01. (D) SKOV3 and SKOV3/DDP cells were pretreated with or without CQ (10 µM) for 2 h and then exposed to the indicated concentrations of cisplatin for 48 h. The cell viability was determined using the Cell Counting Kit-8. The data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 and **P<0.01. LC3, microtubule-associated protein 1 light chain 3; p62, sequestosome 1; GFP, green fluorescent protein; CQ, chloroquine disphosphate.
In order to determine whether cisplatin-induced autophagy exerted a protective function in cisplatin treatment, an autophagy inhibitor, CQ, was used. Treatment with CQ increases the formation of autophagosomes by preventing autophagosome-lysosome fusion, eventually inhibiting late-stage autophagy (27). Cells were treated with cisplatin in the presence or absence of CQ, and cell viability was determined using a CCK8 assay. Cells treated with cisplatin alone were more resistant than those treated with cisplatin and CQ in the examined cell lines (Fig. 2D). These results suggest that cisplatin induces autophagy, serving a protective role in ovarian cancer cells exposed to cisplatin.
Cisplatin promotes BAG3 expression in EOC cells
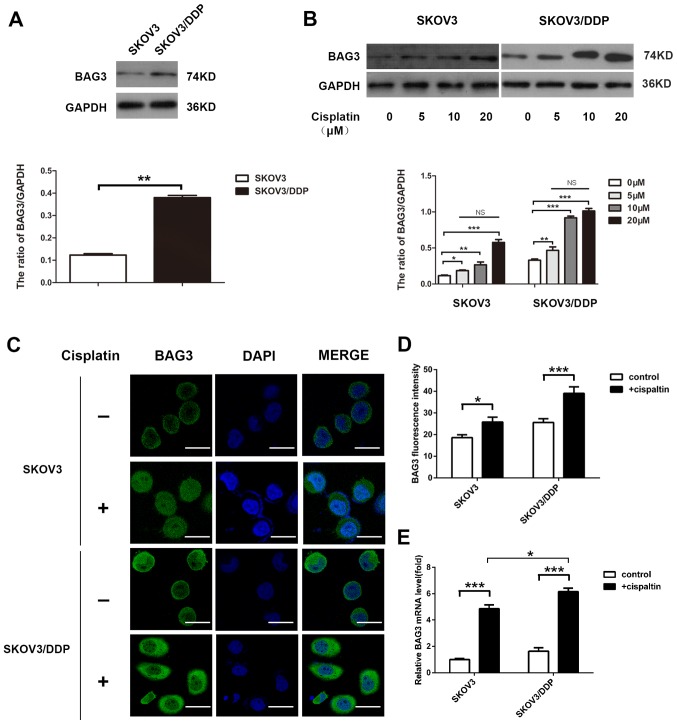
The present study investigated the potential role of BAG3 in the regulation of cisplatin-induced autophagy. The endogenous expression level of BAG3 was lower in the SKOV3 compared with the SKOV3/DDP cells, according to western blot analysis (Fig. 3A). The effect of 48-h treatment with various concentrations of cisplatin on BAG3 expression was then determined. The treatment increased BAG3 protein levels in the cells and the effect was not concentration-dependent. The relative BAG3 expression exhibited no significant differences between different treatment concentration groups (Fig. 3B). Furthermore, confocal microscopy analysis revealed that the green fluorescence of BAG3 accumulated primarily in the cytoplasm, with small amounts in the nucleus (Fig. 3C), and the fluorescence intensity was significantly increased following treatment with 20-µM cisplatin (Fig. 3D). Consistent with the western blotting results, RT-qPCR demonstrated that BAG3 mRNA levels increased following treatment with cisplatin for 48 h, the increase in BAG3 mRNA expression levels in SKOV3/DDP cells was larger than the increase in SKOV3 cells (Fig. 3E).
Figure 3.
Cisplatin promotes the expression of BAG3 in epithelial ovarian cancer cells. (A) SKOV3 and SKOV3/DDP cell lysates were subjected to western blot analysis to determine the endogenous expression level of BAG3 and GAPDH (as a loading control). **P<0.01. (B) SKOV3 and SKOV3/DDP cells were treated with different doses of cisplatin and whole-cell lysates were subjected to western blot analysis to detect BAG3 and GAPDH (as a loading control). Densitometric values were quantified and the data are presented as the mean ± standard deviation of three independent experiments. *P<0.05, **P<0.01 and ***P<0.001 vs. the control group respectively in SKOV3 and SKOV3/DDP. In addition, there are no statistically significant different during different treatment concentration groups respectively in SKOV3 and SKOV3/DDP. (C) SKOV3 and SKOV3/DDP cells were treated with or without 20 µM cisplatin for 48 h and BAG3 expression and location were detected using confocal microscopy. Scale bar, 50 µm. (D) Quantification of the relative fluorescence intensity. The data are presented as the mean ± standard deviation of triplicate determinations. *P<0.05 and ***P<0.001. (E) SKOV3 and SKOV3/DDP cells were treated with or without cisplatin (20 µM) for 48 h and BAG3 mRNA levels were then determined by RT-qPCR. The data are presented as the mean ± standard deviation from three independent experiments. ***P<0.001. BAG3, Bcl-2-associated athanogene 3; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Downregulation of BAG3 modulates cisplatin resistance in ovarian cancer cells via inhibition of autophagy
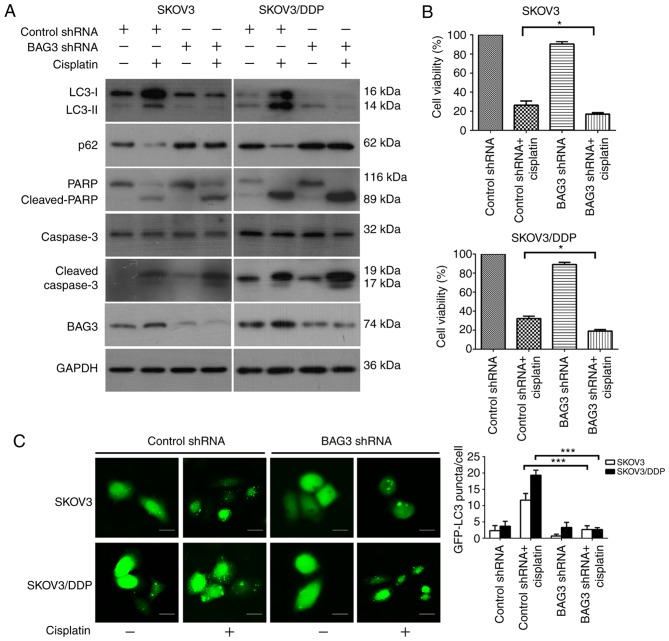
The present study investigated the association between BAG3 and cisplatin-induced autophagy in EOC cells via the knockdown of BAG3 with shRNA. SKOV3/DDP and SKOV3 cells transfected with BAG3 shRNA were treated with cisplatin (20 µM) for 48 h. BAG3 knockdown caused marked decreases in the protein level of BAG3 in the examined cell lines, as demonstrated by western blot analysis. Protein level detection also revealed that the downregulation of BAG3 prevented the formation of LC3-II and the degradation of p62, and enhanced the expression of apoptosis-associated proteins cleaved-PARP and cleaved-caspase-3, following exposure to cisplatin. Furthermore, BAG3 downregulation inhibited autophagy activity more markedly in the SKOV3/DDP cells compared with SKOV3 (Fig. 4A). Subsequently, cell viability was examined using a CCK8 assay. BAG3 knockdown significantly enhanced the sensitivity of the cells to cisplatin compared with that in cells transfected with control shRNA (Fig. 4B). The formation of GFP-LC3 punctate fluorescent dots following treatment with cisplatin were also decreased in SKOV3/DDP cells transfected with BAG3 shRNA, compared with the vector control group (Fig. 4C). Taken together, these results demonstrated that BAG3 serves an important role in the regulation of cisplatin-induced autophagy in ovarian cancer cells.
Figure 4.
Downregulation of BAG3 modulates cisplatin resistance in ovarian cancer cells via the inhibition of autophagy. SKOV3 and SKOV3/DDP cells were transfected with control RNA or shRNA targeting BAG3, followed by treatment with 20 µM cisplatin for 48 h. (A) Whole-cell lysates were subjected to western blot analysis to detect LC3, p62, PARP, cleaved-PARP, caspase-3, cleaved-caspase-3, BAG3 and GAPDH (as a loading control). (B) Cell viability was analyzed using a Cell Counting Kit-8. The data are presented as the mean ± standard deviation from three independent experiments. *P<0.05. (C) SKOV3 and SKOV3/DDP cells were co-transfected with either control or BAG3 shRNA and GFP-LC3 plasmid followed by treatment with cisplatin (20 µM) for an additional 48 h. Scale bar, 200 µm. The formation of GFP-LC3 puncta were examined using immunofluorescence and quantified. The data are presented as the mean ± standard deviation from three independent experiments. ***P<0.001. BAG3, Bcl-2-associated athanogene 3; LC3, microtubule-associated protein 1 light chain 3; p62, sequestosome 1; PARP, poly [ADP-ribose] polymerase; shRNA, short hairpin RNA; GFP, green fluorescent protein.
BAG3 enhances cisplatin-induced apoptosis in EOC cells
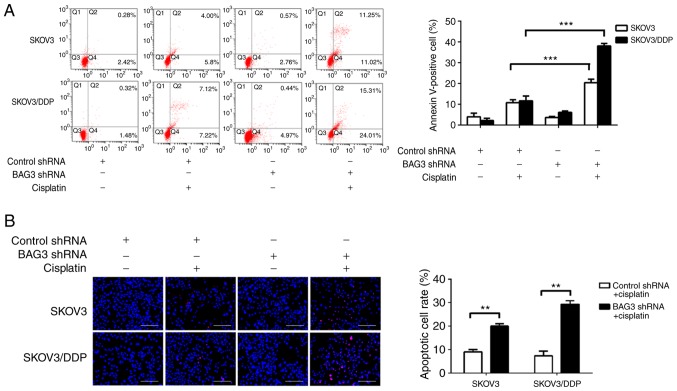
To clarify the role of BAG3 in the regulation of EOC cell apoptosis following cisplatin treatment, shRNA targeting BAG3 was transfected into SKOV3 and SKOV3/DDP cells. Cisplatin induced apoptotic cell death to a great extent after BAG3 knockdown, as revealed by an increase of Annexin V-positive cells by 20.41±1.36 and 38.06±0.99% in SKOV3 and SKOV3/DDP cells, respectively (Fig. 5A). Consistent with the flow cytometry results, the TUNEL assay revealed that cisplatin induced apoptotic cell death to a great degree following BAG3 knockdown in the SKOV3/DDP cells (**P<0.01; Fig. 5B). These data indicate that downregulation of BAG3 enhances cisplatin-induced apoptosis, confirming its role in the resistance of ovarian cancer cell to cisplatin.
Figure 5.
BAG3 inhibits cisplatin-induced apoptosis in epithelial ovarian cancer cells. SKOV3 and SKOV3/DDP cells were transfected with control RNA or shRNA targeting BAG3, followed by treatment with 20 µM cisplatin for 48 h. (A) Apoptosis was analyzed by flow cytometry following Annexin V/propidium iodide staining. The percentages of Annexin V-positive cells are presented in the bar chart. The data are presented as the mean ± SD of three independent experiments. ***P<0.001. (B) Apoptosis was also determined using a TUNEL assay. Scale bar, 400 µm. The percentages of apoptotic cell are presented in the bar chart. **P<0.01. The data are presented as the mean ± SD of three independent experiments. BAG3, Bcl-2-associated athanogene 3; shRNA, short hairpin RNA; SD, standard deviation.
Discussion
The platinum-based cytotoxic compound cisplatin has been commonly used in ovarian cancer treatment for almost three decades, as it induces apoptosis in cancer cells due to lethal DNA damage (28). However, the intrinsic and acquired resistance to cisplatin in cancer cells remains a major challenge. Multiple molecular mechanisms contribute to cisplatin-resistance, including a defective DNA repair system, enhanced drug clearance and detoxification, epigenetic regulation and abnormal signaling pathways (29). Growing evidence indicates a role of autophagy as a pro-survival and resistance mechanism to chemotherapy. A recent study demonstrated that cisplatin can induce autophagy in ovarian cancer cells (5). Herein, a new mechanism is described in which the downregulation of BAG3 mediates cisplatin-induced autophagy in cisplatin-resistant ovarian cancer cells.
Autophagy is an evolutionarily conserved process whereby cytoplasmic cargo sequestered inside double-membrane vesicles is delivered to the lysosome for degradation (30). Autophagy exerts cytoprotective functions and may also lead programmed cell death (31). An increasing amount of evidence suggests that autophagy process may promote cell survival following treatment with anticancer drugs, and may therefore be associated with chemotherapy resistance (32). The induction of autophagy has been demonstrated to promote resistance of ovarian cancer to cisplatin (33). Zhang et al (34) demonstrated that thioredoxin domain containing 17 promoted paclitaxel resistance by inducing autophagy in ovarian cancer. The present study investigated cisplatin resistance in ovarian cancer SKOV3 and SKOV3/DDP cell lines, and the IC50 of cisplatin in the SKOV3/DDP was ~4-fold higher than the parental SKOV3 cells. Cisplatin induced autophagy in a concentration-dependent manner in the examined cells, as demonstrated by the western blot results for the autophagy markers LC3-I, LC3-II and p62. This compound has previously been demonstrated to induce autophagy in various types of cancer cells, such as ovarian cancer cells and osteosarcoma cancer cells (35,36), and another study indicated that autophagy serves a protective role in cisplatin resistance (34). The results of the present study demonstrated that the blockade of autophagy promoted cisplatin-induced cell death in SKOV3 and SKOV3/DDP cells, and partially re-sensitized cisplatin-resistant SKOV3/DDP cells.
BAG3 has been reported to function as a novel modulator of autophagy in cancer cells by regulating key autophagy-related proteins (37). The functions of BAG3 in ovarian cancer have also been partly investigated. A previous study reported that increased BAG3 expression was significantly associated with poor overall survival in patients with primary ovarian tumors (38). BAG3 was also revealed to increase the invasiveness of uterine corpus and ovarian carcinomas (39,40). Furthermore, it also induced resistance to paclitaxel in ovarian clear cell carcinoma cells (41). Recent evidence has suggested that BAG3 exerts a function in adjusting apoptosis and modulating cisplatin resistance (42). The present findings support a role for autophagy activation in chemoresistance in cancer cells, and the downregulation of autophagy was revealed to sensitize the examined cancer cells to cisplatin. Few previous studies have investigated the association between BAG3 and autophagy in ovarian cancer cisplatin resistance, although the current study demonstrated that cisplatin treatment upregulates BAG3 expression. Regulating autophagy to induce cell death, inhibiting protective autophagy, and promoting crosstalk with tissue-specific apoptosis may be promising avenues for novel anticancer chemotherapeutic strategies (43). Therefore, the present study evaluated the role of BAG3 in regulating autophagy in ovarian cancer. The knockdown of BAG3 by shRNA led to the suppression of autophagy, which was measured by a decrease in the level of LC3-II, GFP-LC3 puncta formation and p62 degradation, when compared with the control group, particularly in SKOV3/DDP cells. The downregulation of BAG3 also markedly increased the sensitivity to cisplatin in SKOV3/DDP cells compared with that in SKOV3, as determined by a CCK8 assay. The present study further investigated the association between autophagy and apoptosis with BAG3 downregulation. The knockdown of BAG3 significantly augmented cisplatin-induced apoptosis, as indicated by an increase in the expression of cleaved caspase-3 and PARP. In addition, cisplatin induced apoptotic cell death to a greater degree following BAG3 knockdown, as revealed by Annexin V/PI and TUNEL staining. These results suggest that the downregulation of BAG3 attenuates cisplatin resistance by inhibiting autophagy in ovarian cancer cells. However, the detailed molecular mechanisms underlying the regulation of autophagy via BAG3 may be complex, and further studies are required in order to clarify these.
In conclusion, the present study identified BAG3 as a novel regulator of autophagy and demonstrated its involvement in the modulation of cisplatin resistance in ovarian cancer cells. BAG3 also affected apoptosis, therefore the downregulation of BAG3 can enhance the sensitivity of ovarian cancer cells to cisplatin by regulating autophagy and apoptosis, particularly in the cisplatin-resistant SKOV3/DDP cells. Consequently, BAG3 may represent a novel therapeutic target for preventing chemoresistance in cancer cells, and the knockdown of BAG3 may be a useful strategy to overcome chemoresistance by preventing cytoprotective autophagy in ovarian cancer cells.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- EOC
epithelial ovarian cancer
- BAG3
Bcl-2-associated athanogene 3
- LC3
microtubule-associated protein 1 light chain 3
- p62/SQSTM1
sequestosome 1
- PARP
poly [ADP-ribose] polymerase
- FBS
fetal bovine serum
- CQ
chloroquine disphosphate
- PBS
phosphate-buffered saline
- DMSO
dimethylsulfoxide
- DAPI
4′,6-diamidino-2-phenylindole
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
SQ conceived and designed the study. SQ and LS performed the whole experiment, analyzed the data and wrote the article. YZ and SH analyzed the data and proofread the article.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN, 2012. Int J cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Jeon SY, Hwang KA, Choi KC. Effect of steroid hormones, estrogen and progesterone, on epithelial mesenchymal transition in ovarian cancer development. J Steroid Biochem Mol Biol. 2016;158:1–8. doi: 10.1016/j.jsbmb.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Kim A, Ueda Y, Naka T, Enomoto T. Therapeutic strategies in epithelial ovarian cancer. J Exp Clin Cancer Res. 2012;31:14. doi: 10.1186/1756-9966-31-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang P, Zhang P, Shi B, Zhou M, Jiang H, Zhang H, Pan X, Gao H, Sun H, Li Z. Galectin-1 overexpression promotes progression and chemoresistance to cisplatin in epithelial ovarian cancer. Cell Death Dis. 2014;5:e991. doi: 10.1038/cddis.2013.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Zheng Y, Zhang H, Zhu J, Sun H. Low concentration of chloroquine enhanced efficacy of cisplatin in the treatment of human ovarian cancer dependent on autophagy. Am J Transl Res. 2017;9:4046–4058. [PMC free article] [PubMed] [Google Scholar]
- 6.Maurmann L, Belkacemi L, Adams NR, Majmudar PM, Moghaddas S, Bose RN. A novel cisplatin mediated apoptosis pathway is associated with acid sphingomyelinase and FAS proapoptotic protein activation in ovarian cancer. Apoptosis. 2015;20:960–974. doi: 10.1007/s10495-015-1124-2. [DOI] [PubMed] [Google Scholar]
- 7.Cornelison R, Llaneza DC, Landen CN. Emerging therapeutics to overcome chemoresistance in epithelial ovarian cancer: A mini-review. Int J Mol Sci. 2017;18:E2171. doi: 10.3390/ijms18102171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren F, Shen J, Shi H, Hornicek FJ, Kan Q, Duan Z. Novel mechanisms and approaches to overcome multidrug resistance in the treatment of ovarian cancer. Biochim Biophys Acta. 2016;1866:266–275. doi: 10.1016/j.bbcan.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Lou JS, Bi WC, Chan GKL, Jin Y, Wong CW, Zhou ZY, Wang HY, Yao P, Dong TTX, Tsim KWK. Ginkgetin induces autophagic cell death through p62/SQSTM1-mediated autolysosome formation and redox setting in non-small cell lung cancer. Oncotarget. 2017;8:93131–93148. doi: 10.18632/oncotarget.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naik PP, Mukhopadhyay S, Panda PK, Sinha N, Das CK, Mishra R, Patil S, Bhutia SK. Autophagy regulates cisplatin-induced stemness and chemoresistance via the upregulation of CD44, ABCB1 and ADAM17 in oral squamous cell carcinoma. Cell Prolif. 2018;51 doi: 10.1111/cpr.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pistollato F, Calderón Iglesias R, Ruiz R, Aparicio S, Crespo J, Dzul Lopez L, Giampieri F, Battino M. The use of natural compounds for the targeting and chemoprevention of ovarian cancer. Cancer Lett. 2017;14:191–200. doi: 10.1016/j.canlet.2017.09.050. [DOI] [PubMed] [Google Scholar]
- 12.Zhou WJ, Chang KK, Wu K, Yang HL, Mei J, Xie F, Li DJ, Li MQ. Rapamycin synergizes with cisplatin in antiendometrial cancer activation by improving IL-27-stimulated cytotoxicity of NK cells. Neoplasia. 2018;20:69–79. doi: 10.1016/j.neo.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 14.Wu X, Feng X, Zhao X, Ma F, Liu N, Guo H, Li C, Du H, Zhang B. Role of beclin-1-mediated autophagy in the survival of pediatric leukemia cells. Cell Physiol Biochem. 2016;39:1827–1836. doi: 10.1159/000447882. [DOI] [PubMed] [Google Scholar]
- 15.Liu G, Pei F, Yang F, Li L, Amin AD, Liu S, Buchan JR, Cho WC. Role of autophagy and apoptosis in non-small-cell lung cancer. Int J Mol Sci. 2017;18:E367. doi: 10.3390/ijms18020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behl C. Breaking BAG: The Co-Chaperone BAG3 in health and disease. Trends Pharmacol Sci. 2017;37:672–688. doi: 10.1016/j.tips.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Takayama S, Xie Z, Reed JC. An evolutionarily conserved family of Hsp70/Hsc70 molecular chaperone regulators. J Biol Chem. 1999;274:781–786. doi: 10.1074/jbc.274.2.781. [DOI] [PubMed] [Google Scholar]
- 18.Rauch JN, Tse E, Freilich R, Mok SA, Makley LN, Southworth DR, Gestwicki JE. Bag3 is a modular, scaffolding protein that physically links heat shock protein 70 (Hsp70) to the small heat shock proteins. J Mol Biol. 2017;429:128–141. doi: 10.1016/j.jmb.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knezevic T, Myers VD, Gordon J, Tilley DG, Sharp TE, III, Wang J, Khalili K, Cheung JY, Feldman AM. BAG3: A new player in the heart failure paradigm. Heart Fail Rev. 2015;20:423–434. doi: 10.1007/s10741-015-9487-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, Seguin SJ, Morelli FF, Vinet J, Leo G, et al. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell. 2016;63:796–810. doi: 10.1016/j.molcel.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 21.Felzen V, Hiebel C, Koziollek-Drechsler I, Reißig S, Wolfrum U, Kögel D, Brandts C, Behl C, Morawe T. Estrogen receptor alpha regulates non-canonical autophagy that provides stress resistance to neuroblastoma and breast cancer cells and involves BAG3 function. Cell Death Dis. 2015;6:e1812. doi: 10.1038/cddis.2015.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim EM, Jung CH, Kim J, Hwang SG, Park JK, Um HD. The p53/p21 complex regulates cancer cell invasion and apoptosis by targeting Bcl-2 family proteins. Cancer Res. 2017;77:3092–3100. doi: 10.1158/0008-5472.CAN-16-2098. [DOI] [PubMed] [Google Scholar]
- 23.Chen G, Ke Z, Xu M, Liao M, Wang X, Qi Y, Zhang T, Frank JA, Bower KA, Shi X, Luo J. Autophagy is a protective response to ethanol neurotoxicity. Autophagy. 2012;8:1577–1589. doi: 10.4161/auto.21376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Monitoring autophagy by electron microscopy in mammalian cells. Methods Enzymol. 2009;452:143–164. doi: 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Pan B, Chen D, Huang J, Wang R, Feng B, Song H, Chen L. HMGB1-mediated autophagy promotes docetaxel resistance in human lung adenocarcinoma. Mol Cancer. 2014;13:165. doi: 10.1186/1476-4598-13-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SW, Kim HK, Lee NH, Yi HY, Kim HS, Hong SH, Hong YK, Joe YA. The synergistic effect of combination temozolomide and chloroquine treatment is dependent on autophagy formation and p53 status in glioma cells. Cancer Lett. 2015;360:195–204. doi: 10.1016/j.canlet.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 28.Damia G, Broggini M. Platinum resistance in ovarian cancer: Role of DNA repair. Cancer (Basel) 2019;11:E119. doi: 10.3390/cancers11010119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bookman MA. First-line chemotherapy in epithelial ovarian cancer. Clin Obstet Gynecol. 2012;55:96–113. doi: 10.1097/GRF.0b013e31824b45da. [DOI] [PubMed] [Google Scholar]
- 30.Ahmad L, Mostowy S, Sancho-Shimizu V. Autophagy-virus interplay: From cell biology to human disease. Front Cell Dev Biol. 2018;6:155. doi: 10.3389/fcell.2018.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen N, Karantza V. Autophagy as a therapeutic target in cancer. Cancer Biol Ther. 2011;11:157–168. doi: 10.4161/cbt.11.2.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He C, Dong X, Zhai B, Jiang X, Dong D, Li B, Jiang H, Xu S, Sun X. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;30:28867–28881. doi: 10.18632/oncotarget.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao L, Shi XY, Zhang Y, Zhu Y, Zhu L, Tian W, Zhu BK, Wei ZL. YAP induces cisplatin resistance through activation of autophagy in human ovarian carcinoma cells. Onco Ther. 2016;9:1105–1114. doi: 10.2147/OTT.S102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang SF, Wang XY, Fu ZQ, Peng QH, Zhang JY, Ye F, Fu YF, Zhou CY, Lu WG, Cheng XD, Xie X. TXNDC17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy. 2015;11:225–238. doi: 10.1080/15548627.2014.998931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Jiang W, Hu Y, Da Z, Zeng C, Tu M, Deng Z, Xiao W. MicroRNA-199a-5p inhibits cisplatin-induced drug resistance via inhibition of autophagy in osteosarcoma cells. Oncol Lett. 2016;12:4203–4208. doi: 10.3892/ol.2016.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin WM, Li ZG. Blockage of cisplatin-induced autophagy sensitizes cervical cancer cells to cisplatin. Gene Mol Res. 2015;14:16905–16912. doi: 10.4238/2015.December.14.18. [DOI] [PubMed] [Google Scholar]
- 37.De Marco M, Basile A, Iorio V, Festa M, Falco A, Ranieri B, Pascale M, Sala G, Remodelli P, Capunzo M, et al. Role of BAG3 in cancer progression: A therapeutic opportunity. Semin Cell Dev Biol. 2018;78:85–92. doi: 10.1016/j.semcdb.2017.08.049. [DOI] [PubMed] [Google Scholar]
- 38.Nymoen DA, Hetland Falkenthal TE, Holth A, Ow GS, Ivshina AV, Tropé CG, Kuznetsov VA, Staff AC, Davidson B. Expression and clinical role of chemoresponse-associated genes in ovarian serous carcinoma. Gynecol Oncol. 2015;139:30–39. doi: 10.1016/j.ygyno.2015.07.107. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki M, Iwasaki M, Sugio A, Hishiya A, Tanaka R, Endo T, Takayama S, Saito T. BAG3 (BCL2-associated athanogene 3) interacts with MMP-2 to positively regulate invasion by ovarian carcinoma cells. Cancer Lett. 2011;303:65–71. doi: 10.1016/j.canlet.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 40.Habata S, Iwasaki M, Sugio A, Suzuki M, Tamate M, Satohisa S, Tanaka R, Saito T. BAG3 increases the invasiveness of uterine corpus carcinoma cells by suppressing miR-29b and enhancing MMP2 expression. Oncol Rep. 2015;33:2613–2621. doi: 10.3892/or.2015.3831. [DOI] [PubMed] [Google Scholar]
- 41.Habata S, Iwasaki M, Sugio A, Suzuki M, Tamate M, Satohisa S, Tanaka R, Saito T. BAG3-mediated Mcl-1 stabilization contributes to drug resistance via interaction with USP9X in ovarian cancer. Int J Oncol. 2016;49:402–410. doi: 10.3892/ijo.2016.3494. [DOI] [PubMed] [Google Scholar]
- 42.Wu K, Yang Y, Zhao J, Zhao S. BAG3-mediated miRNA let-7g and let-7i inhibit proliferation and enhance apoptosis of human esophageal carcinoma cells by targeting the drug transporter ABCC10. Cancer Lett. 2016;371:125–133. doi: 10.1016/j.canlet.2015.11.031. [DOI] [PubMed] [Google Scholar]
- 43.Belaid A, Ndiaye PD, Filippakis H, Roux J, Röttinger É, Graba Y, Brest P, Hofman P, Mograbi B. Autophagy: Moving benchside promises to patient bedsides. Curr Cancer Drug Targets. 2015;15:684–702. doi: 10.2174/156800961508151001102452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.