Figure 2.
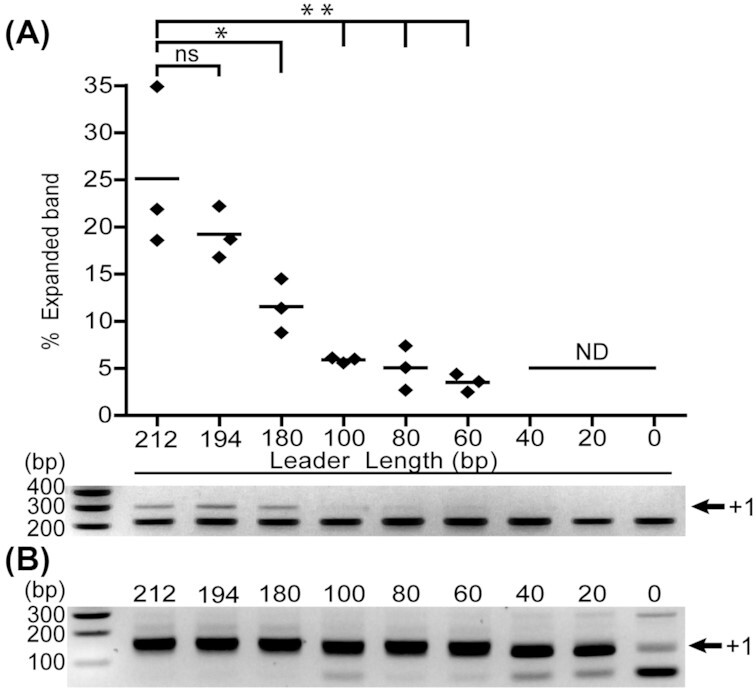
PCR-based detection of spacer acquisition at variable leader length. A—Quantification of expanded CRISPR array band intensity (n = 3). CRISPR adaptation is negatively affected by deletion of motif III that is present in the 212 bp (wild-type) and 194 bp leader (*P < 0.05). Removal of motif II (located in the segment between 180 bp and 100 bp) and motif III significantly reduced (**P < 0.01) acquisition rates close to the detection limit of this PCR. Leaders shorter than 60 bp do not support detectable acquisition (ND). Spacer acquisition rates of the 194 bp leader are not significantly different (ns) from the full 212 bp leader. Statistical significance was calculated using Dunnett's multiple comparisons test. B—Second round of PCR enables the detection of spacer acquisition with leaders shorter than 60 bp or absent leader sequences.
