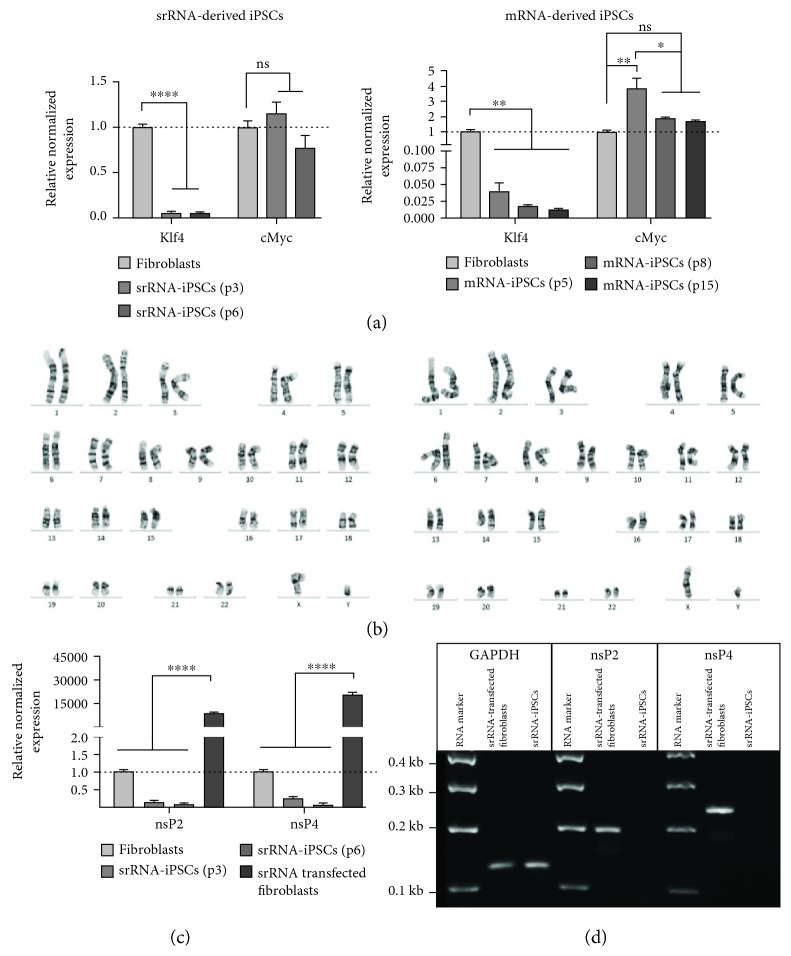
Figure 7.
Analysis of genomic abnormalities, expression of prooncogenic factors, and the presence of srRNA residues in srRNA-iPSCs. (a) Detection of Klf4 and cMyc oncogene expression in iPSCs generated by srRNA and mRNA delivery using qRT-PCR. Results are shown as mean+SEM (n = 3). (b) Representative karyograms of iPSCs generated using srRNA or mRNA. (c) Detection of residual srRNA in obtained srRNA-iPSCs by performing qRT-PCR using nsP2- and nsP4-specific primers. Results are shown as mean+SEM (n = 3). (d) Analysis of specific PCR product lengths using 1% agarose gel electrophoresis. nsP2: 192 bases; nsP4: 238 bases; GAPDH: 126 bases. Statistical differences were determined using one-way ANOVA followed by Bonferroni's multiple comparison test (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001).