Figure 11.
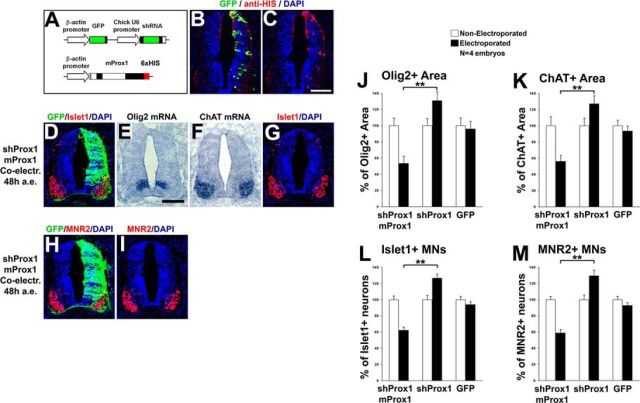
Rescue of the shRNA-Prox1-mediated induction of Olig2 expression and MN generation by murine Prox1. A, Schematic representation of the shProx1 and murine Prox1 expression vectors. Note that shProx1 can be detected with GFP and murine Prox1 is tagged with 6xHIS epitope, and thus can be detected with anti-HIS immunostaining. B, C, Double GFP/HIS immunostaining 48 h after co-electroporation of shProx1 and murine Prox1 constructs. Note that murine Prox1 is distributed in the same cells as the shProx1 construct. D–I, Double immunostainings for GFP/Islet1 (D, G) and GFP/MNR2 (H, I), and in situ hybridizations for Olig2 (E) and ChAT (F), in consecutive sections 48 h a.e., with shProx1 and murine Prox1 constructs. J, K, Quantitative analysis of the Olig2+ (J) and ChAT+ (K) areas presented in E and F, respectively, using ImageJ software. These data are presented as percentage of the nonelectroporated side of the spinal cord. The data for shProx1 electroporations are as in Figure 9F (Olig2) and Figure 10B (ChAT), and are presented here for comparison. For Olig2+ area (E), shProx1 + mProx1 versus shProx1, **p < 0.01 (t test); shProx1 + mProx1 versus GFP, **p < 0.01 (t test), n = 4 embryos. For ChAT+ area (F), shProx1 + mProx1 versus shProx1, **p < 0.01 (t test); shProx1 + mProx1 versus GFP, **p < 0.01 (t test), n = 4 embryos. All cases referred to the electroporated side. L, M, Quantitative analysis of the number of Islet1+ MNs (L) and MNR2+ MNs (M) presented in G and I, respectively. These data are presented as percentage of the nonelectroporated side. The data for shProx1 electroporations are as in Figure 10D (Islet1) and F (MNR2), and presented here for comparison. For Islet+ MNs (L), shProx1 + mProx1 versus shProx1, **p < 0.01 (t test); shProx1 + mProx1 versus GFP, **p < 0.01 (t test), n = 4 embryos. For MNR2+ MNs (M), shProx1 + mProx1 versus shProx1, **p < 0.01 (t test); shProx1 + mProx1 versus GFP, **p < 0.01 (t test), n = 4 embryos. All cases referred to the electroporated side. Scale bars: 100 μm.