Figure 9.
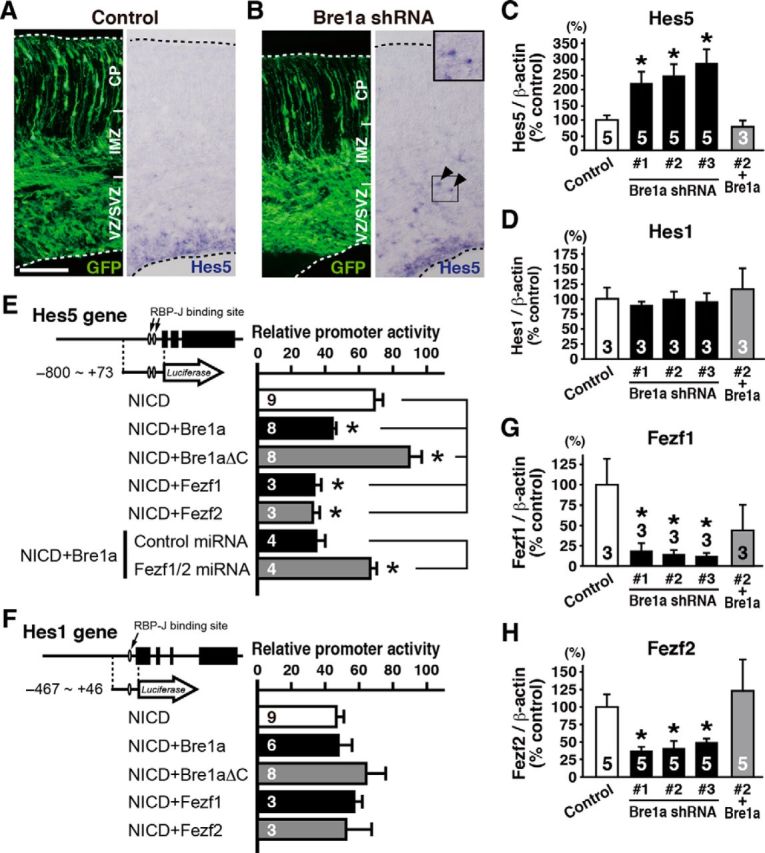
Bre1a knockdown upregulates Hes5 expression in neural precursor cells. A, B, Bre1a or control shRNA and GFP expression plasmids were electroporated into the dorsal forebrain of E13.5 embryos in utero, and brains were analyzed 3 d after the electroporation. Coronal cryosections were immunostained for GFP (left) or subjected to in situ hybridization with an RNA probe for Hes5 (right). Arrowheads indicate ectopic Hes5+ cells outside the VZ. The boxed area is magnified in the inset. Scale bar, 50 μm. C, D, G, H, Primary neurosphere cells derived from E13.5 cortex were infected with lentiviruses expressing Bre1a or control shRNAs and were cultured for 4–7 d. Lentivirus-infected GFP+ neurospheres were used for quantitative RT-PCR analysis of Hes5 (C), Hes1 (D), Fezf1 (G), or Fezf2 (H). For rescue experiments, Bre1a-expressing retrovirus was coinfected with Bre1a shRNA-expressing lentivirus. E, F, Firefly luciferase reporter assays in Neuro2a cells using the following plasmids: a plasmid containing the promoter region of the Hes5 (E) or Hes1 (F) gene; the Renilla luciferase reporter plasmid as an internal control; pCX-NICD, and the indicated plasmid for expression of Bre1a, Bre1aΔC, Fezf1, Fezf2, and Fezf1, and Fezf2 miRNAs. The relative promoter activities were compared with activity in the absence of pCX-NICD. Error bars indicate SEM, and n values are shown in columns. *p < 0.05 by one-way ANOVA followed by Dunnett's post hoc comparison or Student's t test.
