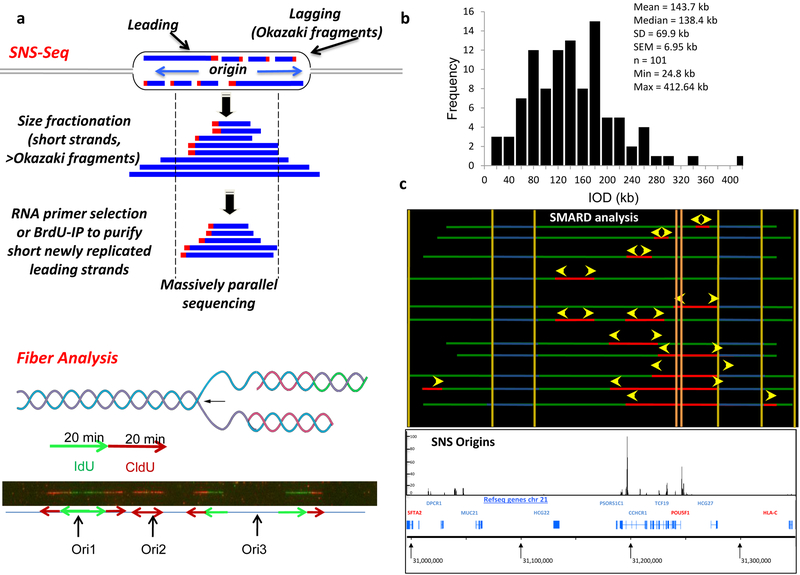
Figure 1: Identification of potential origins in human cells:
(a) The molecular basis of nascent strand sequencing (top) and molecular combing (bottom). Top, DNA replication proceeds bidirectionally from the replication origin (blue arrows). Separation of the two template strands (black) facilitates synthesis of nascent DNA strandss (blue) that proceeds continuously in the 5’ to 3’ directin (leading strand) and discontinuously in the opposite direction (lagging strand – Okazaki fragments). Each nascent DNA strand is primed with an RNA primer (red box). For SNS-Seq, short leading strands are separated based on size fractionation (size range: 400 – 2000 bp to avoid Okazaki fragments) followed by selection of RNA-primed newly replicated DNA based on lambda exonulcease digestion, or selection for DNA that incorporated a nucleotide analog such as BrdU. The population of newly replicated DNA strands is collected, sequenced and aligned ot the refenrece genome. For fiber analysis, DNA is labeled consecutively with two thymidine analogs (e.g. CldU and IdU) during DNA synthesis. DNA is isolated, stretched in microscope slides and visualized following detection with specific antibodies that recognize the nucleotide analogs. Replication origins are identified as regions of green flanked by red (ori1), red without green (ori2) or regions that are flanked first by green and then by red (ori3). (b) Histogram showing the distribution of inter-origin distances (IOD) measured by molecular combing in IMR-90 cells. (c) Comparison of Single Molecule Analysis of Replicating DNA (SMARD) analysis on the region containing the POU5F1 locus region (chr6:30,996,000–31,350,000) in hESC cells (top panel - reproduced from Desprat et al. 2009 Genome Res.) with significant origin peaks identified with Sole-Search in IMR-90 cells (bottom panel) (enrichments of SNS are shown in IGB without background).