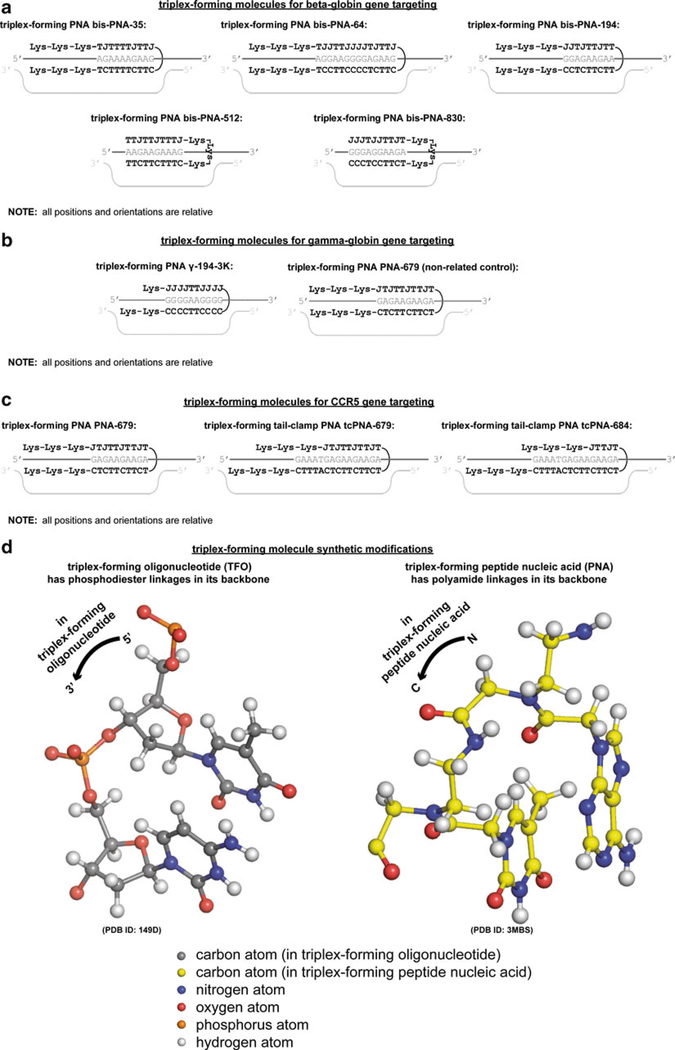
Fig. 2.
(continued) Synthetic mutagenic triplex-forming molecules and their genomic targeting sites. Triplex-forming molecules have been designed to target genomic sites, thereby upregulating homologous recombination with the recombinagenic donor DNA molecules, or as non-related controls, to examine targeting specificity, for (a) the beta-globin gene, where triplex-forming bis-PNAs are used, (b) the gamma-globin gene, where triplex-forming PNAs were used, (c) and the CCR5 gene, where triplex-forming PNAs and triplex-forming tail-clamp PNAs were used. (d) For these exogenously introduced triplex-forming molecules, positive design is implemented based on major groove binding Hoogsteen oriented base pairing and strand-invading Watson-Crick oriented base pairing with genomic DNA, as well as negative design based on synthetic modifications to the linkages in the backbone to prevent endogenous nuclease and protease-mediated degradation and enhance electrostatic complementarity to negatively charged genomic DNA. Linkages for triplex-forming oligonucleotide and triplex-forming peptide nucleic acid molecular modeled from [98] and [99], respectively