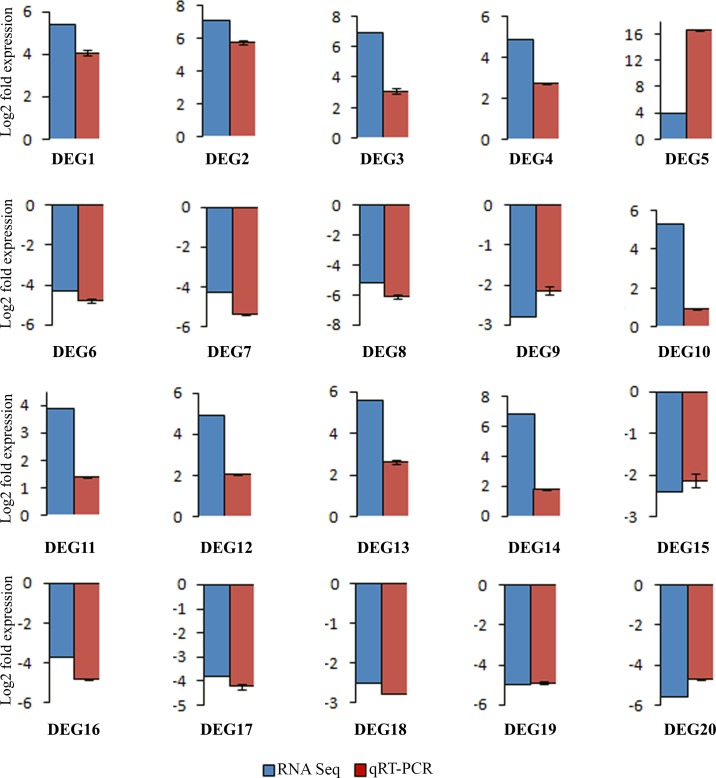
Fig 8.
Validation and comparison of the RNA-seq and qRT- PCR expression profile of differentially expressed genes [Cysteine- rich receptor- like protein 3 (DEG1); G-type lectin receptor-like serine/threonine protein kinase (DEG2); Receptor-like protein kinase FERONIA (DEG3); Wall-associated receptor kinase-like 14 (DEG4); LysM domain receptor-like kinase 3 (DEG5); L-type lectin-domain containing receptor kinase VIII.2-like (DEG6); Transcription factor PIF3-like (DEG7); Heat stress transcription factor A-6b-like (DEG8); Protein LHY isoform X3 (DEG9); Transcription factor bHLH48-like (DEG10); calmodulin-binding transcription activator 1-like isoform X2 (DEG11); Probable WRKY transcription factor 41 (DEG12); probable methyltransferase 19 (DEG13); Subtilisin-like protease SBT1.6 (DEG14); U-box domain-containing protein 4 (DEG15); Zeatin expoxidase (DEG16); Delta-1-pyrroline-5-carboxylate synthase-like isoform X2 (DEG17); Flavonol synthase/flavanone 3-hydroxylase (DEG18); Probable inositol transporter 2 isoform X2 (DEG19); B-box zinc finger protein 18-like (DEG20)].