Fig. 4. CATACOMB and PRC2 catalytic activity are required for U2OS proliferation.
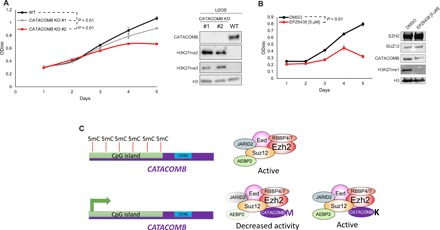
(A) Left: Growth curve comparison between U2OS WT parental cells and U2OS CATACOMB KO clones. Absorbance of crystal violet in 10% acetic acid was used to represent the cell number and plotted as optical density (OD) (λ = 590). n = 3. Error bars represent SDs. Right: Western blotting of total cell extracts from cells described in the left part of the panel. Antibodies used for Western blotting are indicated in the panel. H3 served as a loading control. P values were calculated using the areas under the curves. (B) Left: Growth curve comparison between U2OS cells treated with 5 μM EPZ6438 (EZH2 inhibitor) or DMSO (vehicle control). Cells were pretreated for 6 days, every other day for 6 days, and kept on the same regimen during the 5 days that the proliferation was assessed. Absorbance of crystal violet in 10% acetic acid was used to represent the cell number and plotted as OD (λ = 590). n = 3. Error bars represent SDs. Right: Western blotting of total cell extracts from cells shown in the left panel. Antibodies used for Western blotting are indicated in the panel. H3 served as a loading control. (C) Working model proposed by our study. CATACOMB is kept silenced by DNA methylation; once demethylation occurs, the gene is activated, the protein product binds to the PRC2, and through the conserved M406 residue decreases PRC2 activity. Single methionine-to-lysine mutation (M406K) abolishes CATACOMB inhibitory activity toward PRC2 without disrupting the binding between CATACOMB and PRC2.
